A flexible loop in the paxillin LIM3 domain mediates its direct binding to integrin β subunits
- PMID: 39231388
- PMCID: PMC11374337
- DOI: 10.1371/journal.pbio.3002757
A flexible loop in the paxillin LIM3 domain mediates its direct binding to integrin β subunits
Abstract
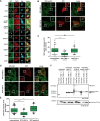
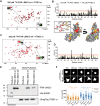
Integrins are fundamental for cell adhesion and the formation of focal adhesions (FA). Accordingly, these receptors guide embryonic development, tissue maintenance, and haemostasis but are also involved in cancer invasion and metastasis. A detailed understanding of the molecular interactions that drive integrin activation, FA assembly, and downstream signalling cascades is critical. Here, we reveal a direct association of paxillin, a marker protein of FA sites, with the cytoplasmic tails of the integrin β1 and β3 subunits. The binding interface resides in paxillin's LIM3 domain, where based on the NMR structure and functional analyses, a flexible, 7-amino acid loop engages the unstructured part of the integrin cytoplasmic tail. Genetic manipulation of the involved residues in either paxillin or integrin β3 compromises cell adhesion and motility of murine fibroblasts. This direct interaction between paxillin and the integrin cytoplasmic domain identifies an alternative, kindlin-independent mode of integrin outside-in signalling particularly important for integrin β3 function.
Copyright: © 2024 Baade et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

