Heterozygous RPA2 variant as a novel genetic cause of telomere biology disorders
- PMID: 39231615
- PMCID: PMC11444173
- DOI: 10.1101/gad.352032.124
Heterozygous RPA2 variant as a novel genetic cause of telomere biology disorders
Abstract
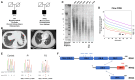
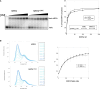
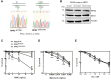
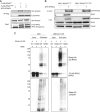
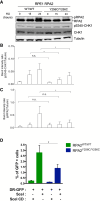
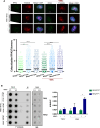
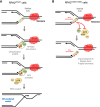
Premature telomere shortening or telomere instability is associated with a group of rare and heterogeneous diseases collectively known as telomere biology disorders (TBDs). Here we identified two unrelated individuals with clinical manifestations of TBDs and short telomeres associated with the identical monoallelic variant c.767A>G; Y256C in RPA2 Although the replication protein A2 (RPA2) mutant did not affect ssDNA binding and G-quadruplex-unfolding properties of RPA, the mutation reduced the affinity of RPA2 with the ubiquitin ligase RFWD3 and reduced RPA ubiquitination. Using engineered knock-in cell lines, we found an accumulation of RPA at telomeres that did not trigger ATR activation but caused short and dysfunctional telomeres. Finally, both patients acquired, in a subset of blood cells, somatic genetic rescue events in either POT1 genes or TERT promoters known to counteract the accelerated telomere shortening. Collectively, our study indicates that variants in RPA2 represent a novel genetic cause of TBDs. Our results further support the fundamental role of the RPA complex in regulating telomere length and stability in humans.
Keywords: RFWD3; RPA; telomere; telomere disorders; telomere replication.
© 2024 Kochman et al.; Published by Cold Spring Harbor Laboratory Press.
Figures

Similar articles
-
RPA and POT1: friends or foes at telomeres?Cell Cycle. 2012 Feb 15;11(4):652-7. doi: 10.4161/cc.11.4.19061. Cell Cycle. 2012. PMID: 22373525 Free PMC article.
-
Characterization of novel mutations in the TEL-patch domain of the telomeric factor TPP1 associated with telomere biology disorders.Hum Mol Genet. 2024 Mar 20;33(7):612-623. doi: 10.1093/hmg/ddad210. Hum Mol Genet. 2024. PMID: 38176734
-
RPA prevents G-rich structure formation at lagging-strand telomeres to allow maintenance of chromosome ends.EMBO J. 2015 Jul 14;34(14):1942-58. doi: 10.15252/embj.201490773. Epub 2015 Jun 3. EMBO J. 2015. PMID: 26041456 Free PMC article.
-
POT1 mutations cause differential effects on telomere length leading to opposing disease phenotypes.J Cell Physiol. 2023 Jun;238(6):1237-1255. doi: 10.1002/jcp.31034. Epub 2023 May 14. J Cell Physiol. 2023. PMID: 37183325 Review.
-
Structural biology of telomeres and telomerase.Cell Mol Life Sci. 2020 Jan;77(1):61-79. doi: 10.1007/s00018-019-03369-x. Epub 2019 Nov 14. Cell Mol Life Sci. 2020. PMID: 31728577 Free PMC article. Review.
Cited by
-
Insights into the length and breadth of methodologies harnessed to study human telomeres.Biomark Res. 2024 Oct 22;12(1):127. doi: 10.1186/s40364-024-00668-9. Biomark Res. 2024. PMID: 39438947 Free PMC article.
References
-
- Bertrand A, Ba I, Kermasson L, Pirabakaran V, Chable N, Lainey E, Ménard C, Kallel F, Picard C, Hadiji S, et al. 2024. Characterization of novel mutations in the TEL-patch domain of the telomeric factor TPP1 associated with telomere biology disorders. Hum Mol Genet 33: 612–623. 10.1093/hmg/ddad210 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
