G protein selectivity profile of GPR56/ADGRG1 and its effect on downstream effectors
- PMID: 39231834
- PMCID: PMC11374949
- DOI: 10.1007/s00018-024-05416-8
G protein selectivity profile of GPR56/ADGRG1 and its effect on downstream effectors
Abstract
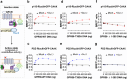
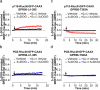
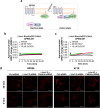
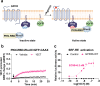
GPR56, an adhesion G-protein coupled receptor (aGPCRs) with constitutive and ligand-promoted activity, is involved in many physiological and pathological processes. Whether the receptor's constitutive or ligand-promoted activation occur through the same molecular mechanism, and whether different activation modes lead to functional selectivity between G proteins is unknown. Here we show that GPR56 constitutively activates both G12 and G13. Unlike constitutive activation and activation with 3-α-acetoxydihydrodeoxygedunin (3αDOG), stimulation with an antibody, 10C7, directed against GPR56's extracellular domain (ECD) led to an activation that favors G13 over G12. An autoproteolytically deficient mutant, GPR56-T383A, was also activated by 10C7 indicating that the tethered agonist (TA) exposed through autocatalytic cleavage, is not required for this activation modality. In contrast, this proteolysis-resistant mutant could not be activated by 3αDOG indicating different modes of activation by the two ligands. We show that an N-terminal truncated GPR56 construct (GPR56-Δ1-385) is devoid of constitutive activity but was activated by 3αDOG. Similarly to 3αDOG, 10C7 promoted the recruitment of β-arrestin-2 but GPR56 internalization was β-arrestin independent. Despite the slow activation mode of 10C7 that favors G13 over G12, it efficiently activated the downstream Rho pathway in BT-20 breast cancer cells. These data show that different GPR56 ligands have different modes of activation yielding differential G protein selectivity but converging on the activation of the Rho pathway both in heterologous expressions system and in cancer cells endogenously expressing the receptor. 10C7 is therefore an interesting tool to study both the processes underlying GPR56 activity and its role in cancer cells.
Keywords: BRET; G protein-coupled receptor; GPR56; RhoGEF; Signaling; Trafficking.
© 2024. The Author(s).
Conflict of interest statement
Some of the BRET biosensors used in the present study are protected by patents and are licensed to Domain Therapeutics for commercial use. MB is the president of Domain Therapeutics scientific advisory board. All other authors declare no conflict of interest.
Figures


Update of
-
G protein selectivity profile of GPR56/ADGRG1 and its effect on downstream effectors.Res Sq [Preprint]. 2024 Sep 5:rs.3.rs-4869264. doi: 10.21203/rs.3.rs-4869264/v1. Res Sq. 2024. Update in: Cell Mol Life Sci. 2024 Sep 4;81(1):383. doi: 10.1007/s00018-024-05416-8. PMID: 39281883 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

