Ingestion of Bacillus cereus spores dampens the immune response to favor bacterial persistence
- PMID: 39231950
- PMCID: PMC11375157
- DOI: 10.1038/s41467-024-51689-9
Ingestion of Bacillus cereus spores dampens the immune response to favor bacterial persistence
Abstract
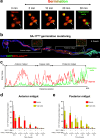
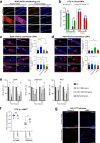
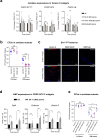
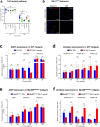
Strains of the Bacillus cereus (Bc) group are sporulating bacteria commonly associated with foodborne outbreaks. Spores are dormant cells highly resistant to extreme conditions. Nevertheless, the pathological processes associated with the ingestion of either vegetative cells or spores remain poorly understood. Here, we demonstrate that while ingestion of vegetative bacteria leads to their rapid elimination from the intestine of Drosophila melanogaster, a single ingestion of spores leads to the persistence of bacteria for at least 10 days. We show that spores do not germinate in the anterior part of the intestine which bears the innate immune defenses. Consequently, spores reach the posterior intestine where they germinate and activate both the Imd and Toll immune pathways. Unexpectedly, this leads to the induction of amidases, which are negative regulators of the immune response, but not to antimicrobial peptides. Thereby, the local germination of spores in the posterior intestine dampens the immune signaling that in turn fosters the persistence of Bc bacteria. This study provides evidence for how Bc spores hijack the intestinal immune defenses allowing the localized birth of vegetative bacteria responsible for the digestive symptoms associated with foodborne illness outbreaks.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Revisiting bacterial spore germination in the presence of peptidoglycan fragments.J Bacteriol. 2025 Jul 24;207(7):e0014625. doi: 10.1128/jb.00146-25. Epub 2025 Jul 3. J Bacteriol. 2025. PMID: 40607799 Free PMC article.
-
Sporulation at reduced water activity impairs germination kinetics of Bacillus subtilis spores.Appl Environ Microbiol. 2025 Jul 23;91(7):e0067725. doi: 10.1128/aem.00677-25. Epub 2025 Jun 10. Appl Environ Microbiol. 2025. PMID: 40492716 Free PMC article.
-
Drosophila melanogaster Toll-9 elicits antiviral immunity against Drosophila C virus.J Virol. 2025 Jun 17;99(6):e0221424. doi: 10.1128/jvi.02214-24. Epub 2025 May 14. J Virol. 2025. PMID: 40366172 Free PMC article.
-
How lived experiences of illness trajectories, burdens of treatment, and social inequalities shape service user and caregiver participation in health and social care: a theory-informed qualitative evidence synthesis.Health Soc Care Deliv Res. 2025 Jun;13(24):1-120. doi: 10.3310/HGTQ8159. Health Soc Care Deliv Res. 2025. PMID: 40548558
-
Modulation of immunity and inflammatory gene expression in the gut, in inflammatory diseases of the gut and in the liver by probiotics.World J Gastroenterol. 2014 Nov 14;20(42):15632-49. doi: 10.3748/wjg.v20.i42.15632. World J Gastroenterol. 2014. PMID: 25400447 Free PMC article.
Cited by
-
Update of the list of qualified presumption of safety (QPS) recommended microbiological agents intentionally added to food or feed as notified to EFSA 21: Suitability of taxonomic units notified to EFSA until September 2024.EFSA J. 2025 Jan 20;23(1):e9169. doi: 10.2903/j.efsa.2025.9169. eCollection 2025 Jan. EFSA J. 2025. PMID: 39834754 Free PMC article.
-
Spatial and temporal coordination of Duox/TrpA1/Dh31 and IMD pathways is required for the efficient elimination of pathogenic bacteria in the intestine of Drosophila larvae.Elife. 2024 Nov 22;13:RP98716. doi: 10.7554/eLife.98716. Elife. 2024. PMID: 39576741 Free PMC article.
-
New Approach Methods to Assess the Enteropathogenic Potential of Strains of the Bacillus cereus Group, including Bacillus thuringiensis.Foods. 2024 Apr 9;13(8):1140. doi: 10.3390/foods13081140. Foods. 2024. PMID: 38672813 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- ANR-15-IDEX-01/Agence Nationale de la Recherche (French National Research Agency)
- ANR-13-CESA-0003-01/Agence Nationale de la Recherche (French National Research Agency)
- ANR-22-CE35-0006-01/Agence Nationale de la Recherche (French National Research Agency)
- 20171206145/Fondation ARC pour la Recherche sur le Cancer (ARC Foundation for Cancer Research)
- PhD half-grant/Institut National de la Recherche Agronomique (National Institute for Agricultural Research)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

