Human milk oligosaccharides are associated with maternal genetics and respiratory health of human milk-fed children
- PMID: 39232002
- PMCID: PMC11375010
- DOI: 10.1038/s41467-024-51743-6
Human milk oligosaccharides are associated with maternal genetics and respiratory health of human milk-fed children
Abstract
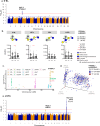
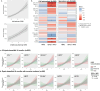
Breastfeeding provides many health benefits, but its impact on respiratory health remains unclear. This study addresses the complex and dynamic nature of the mother-milk-infant triad by investigating maternal genomic factors regulating human milk oligosaccharides (HMOs), and their associations with respiratory health among human milk-fed infants. Nineteen HMOs are quantified from 980 mothers of the CHILD Cohort Study. Genome-wide association studies identify HMO-associated loci on chromosome 19p13.3 and 19q13.33 (lowest P = 2.4e-118), spanning several fucosyltransferase (FUT) genes. We identify novel associations on chromosome 3q27.3 for 6'-sialyllactose (P = 2.2e-9) in the sialyltransferase (ST6GAL1) gene. These, plus additional associations on chromosomes 7q21.32, 7q31.32 and 13q33.3, are replicated in the independent INSPIRE Cohort. Moreover, gene-environment interaction analyses suggest that fucosylated HMOs may modulate overall risk of recurrent wheeze among preschoolers with variable genetic risk scores (P < 0.01). Thus, we report novel genetic factors associated with HMOs, some of which may protect the respiratory health of children.
© 2024. The Author(s).
Conflict of interest statement
J.C. is currently an employee of F. Hoffman-La Roche Ltd., however, the published work was done prior to this employment and does not involve/promote any of Roche’s materials or point of view. M.B.A. has consulted for DSM Nutritional Products (a food ingredient company) and serves on the Scientific Advisory Board for TinyHealth (a microbiome testing company). She has received research funding (unrelated to this project) and speaking honoraria from Prolacta Biosciences (a huma milk fortifier company). L.B. is a co-inventor on patent applications related to the use of HMOs in preventing NEC and other inflammatory diseases. The remaining authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

