Differential effects of 40S ribosome recycling factors on reinitiation at regulatory uORFs in GCN4 mRNA are not dictated by their roles in bulk 40S recycling
- PMID: 39232119
- PMCID: PMC11375166
- DOI: 10.1038/s42003-024-06761-x
Differential effects of 40S ribosome recycling factors on reinitiation at regulatory uORFs in GCN4 mRNA are not dictated by their roles in bulk 40S recycling
Abstract
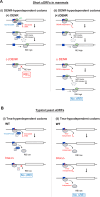
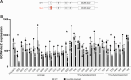
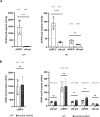
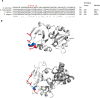
Recycling of 40S ribosomal subunits following translation termination, entailing release of deacylated tRNA and dissociation of the empty 40S from mRNA, involves yeast Tma20/Tma22 heterodimer and Tma64, counterparts of mammalian MCTS1/DENR and eIF2D. MCTS1/DENR enhance reinitiation (REI) at short upstream open reading frames (uORFs) harboring penultimate codons that confer heightened dependence on these factors in bulk 40S recycling. Tma factors, by contrast, inhibited REI at particular uORFs in extracts; however, their roles at regulatory uORFs in vivo were unknown. We examined effects of eliminating Tma proteins on REI at regulatory uORFs mediating translational control of GCN4 optimized for either promoting (uORF1) or preventing (uORF4) REI. We found that the Tma proteins generally impede REI at native uORF4 and its variants equipped with various penultimate codons regardless of their Tma-dependence in bulk recycling. The Tma factors have no effect on REI at native uORF1 and equipping it with Tma-hyperdependent penultimate codons generally did not confer Tma-dependent REI; nor did converting the uORFs to AUG-stop elements. Thus, effects of the Tma proteins vary depending on the REI potential of the uORF and penultimate codon, but unlike in mammals, are not principally dictated by the Tma-dependence of the codon in bulk 40S recycling.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

