Structure of tetrameric forms of the serotonin-gated 5-HT3A receptor ion channel
- PMID: 39232129
- PMCID: PMC11480441
- DOI: 10.1038/s44318-024-00191-5
Structure of tetrameric forms of the serotonin-gated 5-HT3A receptor ion channel
Abstract
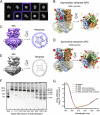
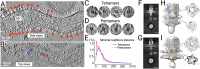
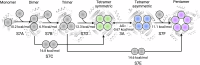
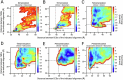
Multimeric membrane proteins are produced in the endoplasmic reticulum and transported to their target membranes which, for ion channels, is typically the plasma membrane. Despite the availability of many fully assembled channel structures, our understanding of assembly intermediates, multimer assembly mechanisms, and potential functions of non-standard assemblies is limited. We demonstrate that the pentameric ligand-gated serotonin 5-HT3A receptor (5-HT3AR) can assemble to tetrameric forms and report the structures of the tetramers in plasma membranes of cell-derived microvesicles and in membrane memetics using cryo-electron microscopy and tomography. The tetrameric structures have near-symmetric transmembrane domains, and asymmetric extracellular domains, and can bind serotonin molecules. Computer simulations, based on our cryo-EM structures, were used to decipher the assembly pathway of pentameric 5-HT3R and suggest a potential functional role for the tetrameric receptors.
Keywords: Cryo-EM; Ion Channels; Pentameric Ligand-gated Ion Channels; Serotonin Receptors.
© 2024. The Author(s).
Figures








References
-
- Barducci A, Bonomi M, Parrinello M (2011) Metadynamics. WIREs Comput Mol Sci 1:826–843
-
- Barducci A, Bussi G, Parrinello M (2008) Well-tempered metadynamics: a smoothly converging and tunable free-energy method. Phys Rev Lett 100:020603 - PubMed
-
- Barrera NP, Henderson RM, Edwardson JM (2008) Determination of the architecture of ionotropic receptors using AFM imaging. Pflüg Arch - Eur J Physiol 456:199–209 - PubMed
-
- Bas DC, Rogers DM, Jensen JH (2008) Very fast prediction and rationalization of pKa values for protein–ligand complexes. Proteins Struct Funct Bioinforma 73:765–783 - PubMed
MeSH terms
Substances
Grants and funding
- KU3222/2-1,KU3222/3-1/Deutsche Forschungsgemeinschaft (DFG)
- JCYJ20200109114818703/CAS | Shenzhen Institutes of Advanced Technology, Chinese Academy of Sciences (SIAT, CAS)
- 2019QN01Y306/Guangdong province
- Academician Work funding/Guangdong province
- ZDSYS20201230165400001/Shenzhen Key Laboratory for Computer Aided Drug Discovery in SIAT
LinkOut - more resources
Full Text Sources

