CDK5-cyclin B1 regulates mitotic fidelity
- PMID: 39232161
- PMCID: PMC12307448
- DOI: 10.1038/s41586-024-07888-x
CDK5-cyclin B1 regulates mitotic fidelity
Abstract
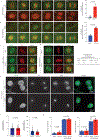
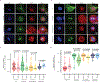
CDK1 has been known to be the sole cyclin-dependent kinase (CDK) partner of cyclin B1 to drive mitotic progression1. Here we demonstrate that CDK5 is active during mitosis and is necessary for maintaining mitotic fidelity. CDK5 is an atypical CDK owing to its high expression in post-mitotic neurons and activation by non-cyclin proteins p35 and p392. Here, using independent chemical genetic approaches, we specifically abrogated CDK5 activity during mitosis, and observed mitotic defects, nuclear atypia and substantial alterations in the mitotic phosphoproteome. Notably, cyclin B1 is a mitotic co-factor of CDK5. Computational modelling, comparison with experimentally derived structures of CDK-cyclin complexes and validation with mutational analysis indicate that CDK5-cyclin B1 can form a functional complex. Disruption of the CDK5-cyclin B1 complex phenocopies CDK5 abrogation in mitosis. Together, our results demonstrate that cyclin B1 partners with both CDK5 and CDK1, and CDK5-cyclin B1 functions as a canonical CDK-cyclin complex to ensure mitotic fidelity.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Figures















References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

