A protein risk score for all-cause and respiratory-specific mortality in non-Hispanic white and African American individuals who smoke
- PMID: 39232179
- PMCID: PMC11374806
- DOI: 10.1038/s41598-024-71714-7
A protein risk score for all-cause and respiratory-specific mortality in non-Hispanic white and African American individuals who smoke
Abstract
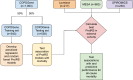
Protein biomarkers are associated with mortality in cardiovascular disease, but their effect on predicting respiratory and all-cause mortality is not clear. We tested whether a protein risk score (protRS) can improve prediction of all-cause mortality over clinical risk factors in smokers. We utilized smoking-enriched (COPDGene, LSC, SPIROMICS) and general population-based (MESA) cohorts with SomaScan proteomic and mortality data. We split COPDGene into training and testing sets (50:50) and developed a protRS based on respiratory mortality effect size and parsimony. We tested multivariable associations of the protRS with all-cause, respiratory, and cardiovascular mortality, and performed meta-analysis, area-under-the-curve (AUC), and network analyses. We included 2232 participants. In COPDGene, a penalized regression-based protRS was most highly associated with respiratory mortality (OR 9.2) and parsimonious (15 proteins). This protRS was associated with all-cause mortality (random effects HR 1.79 [95% CI 1.31-2.43]). Adding the protRS to clinical covariates improved all-cause mortality prediction in COPDGene (AUC 0.87 vs 0.82) and SPIROMICS (0.74 vs 0.6), but not in LSC and MESA. Protein-protein interaction network analyses implicate cytokine signaling, innate immune responses, and extracellular matrix turnover. A blood-based protein risk score predicts all-cause and respiratory mortality, identifies potential drivers of mortality, and demonstrates heterogeneity in effects amongst cohorts.
© 2024. This is a U.S. Government work and not under copyright protection in the US; foreign copyright protection may apply.
Conflict of interest statement
E.K.S. received grant support from Northpond Laboratories and Bayer. M.H.C. has received grant support from Bayer. M.M. received grant support from Bayer and consulting fees from Sitka, TheaHealth, 2ndMD, TriNetX, Verona Pharma, and Axon Advisors.
Figures


References
MeSH terms
Substances
Grants and funding
- R01HL137927/HL/NHLBI NIH HHS/United States
- R01 HL153248/HL/NHLBI NIH HHS/United States
- U01 HL089856/HL/NHLBI NIH HHS/United States
- R01 HL135142/HL/NHLBI NIH HHS/United States
- R01HL153248/HL/NHLBI NIH HHS/United States
- K08HL159318/HL/NHLBI NIH HHS/United States
- R01 HL147148/HL/NHLBI NIH HHS/United States
- R01 HL089856/HL/NHLBI NIH HHS/United States
- U01 HL137880/HL/NHLBI NIH HHS/United States
- HL089856/HL/NHLBI NIH HHS/United States
- K08 HL159318/HL/NHLBI NIH HHS/United States
- R01 HL137927/HL/NHLBI NIH HHS/United States
- HL147148/HL/NHLBI NIH HHS/United States
- R01HL135142/HL/NHLBI NIH HHS/United States

