An hepatitis B and D virus infection model using human pluripotent stem cell-derived hepatocytes
- PMID: 39232200
- PMCID: PMC11466959
- DOI: 10.1038/s44319-024-00236-0
An hepatitis B and D virus infection model using human pluripotent stem cell-derived hepatocytes
Abstract
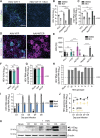
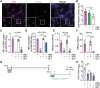
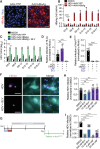
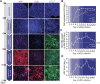
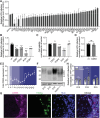
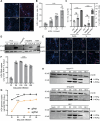
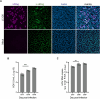
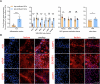
Current culture systems available for studying hepatitis D virus (HDV) are suboptimal. In this study, we demonstrate that hepatocyte-like cells (HLCs) derived from human pluripotent stem cells (hPSCs) are fully permissive to HDV infection across various tested genotypes. When co-infected with the helper hepatitis B virus (HBV) or transduced to express the HBV envelope protein HBsAg, HLCs effectively release infectious progeny virions. We also show that HBsAg-expressing HLCs support the extracellular spread of HDV, thus providing a valuable platform for testing available anti-HDV regimens. By challenging the cells along the differentiation with HDV infection, we have identified CD63 as a potential HDV co-entry factor that was rate-limiting for HDV infection in immature hepatocytes. Given their renewable source and the potential to derive hPSCs from individual patients, we propose HLCs as a promising model for investigating HDV biology. Our findings offer new insights into HDV infection and expand the repertoire of research tools available for the development of therapeutic interventions.
Keywords: Antiviral Treatment; Hepatitis B Virus; Hepatitis D Virus; Hepatocyte-like Cells; Human Pluripotent Stem Cells.
© 2024. The Author(s).
Conflict of interest statement
Stephan Urban is a co-inventor and applicant on patents protecting HBV pre-S1-derived lipopeptides (Myrcludex B/Bulevirtide/Hepcludex). The remaining authors declare no competing interests.
Figures


References
-
- Alexa A, Rahnenfuhrer J (2016) topGO: enrichment analysis for Gene Ontology. R Packag. version 2.26.0. R Package version 2260
-
- Alfaiate D, Dény P, Durantel D (2015) Hepatitis delta virus: from biological and medical aspects to current and investigational therapeutic options. Antivir Res 122:112–129 - PubMed
-
- Appelman MD, Wettengel JM, Protzer U, Oude Elferink RPJ, van de Graaf SFJ (2021) Molecular regulation of the hepatic bile acid uptake transporter and HBV entry receptor NTCP. Biochimica et Biophysica Acta - Mol Cell Biol Lipids 1866:158960 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

