Inflammation-induced epigenetic imprinting regulates intestinal stem cells
- PMID: 39232559
- PMCID: PMC11963838
- DOI: 10.1016/j.stem.2024.08.006
Inflammation-induced epigenetic imprinting regulates intestinal stem cells
Abstract
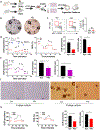
It remains unknown whether and how intestinal stem cells (ISCs) adapt to inflammatory exposure and whether the adaptation leaves scars that will affect their subsequent regeneration. We investigated the consequences of inflammation on Lgr5+ ISCs in well-defined clinically relevant models of acute gastrointestinal graft-versus-host disease (GI GVHD). Utilizing single-cell transcriptomics, as well as organoid, metabolic, epigenomic, and in vivo models, we found that Lgr5+ ISCs undergo metabolic changes that lead to the accumulation of succinate, which reprograms their epigenome. These changes reduced the ability of ISCs to differentiate and regenerate ex vivo in serial organoid cultures and also in vivo following serial transplantation. Furthermore, ISCs demonstrated a reduced capacity for in vivo regeneration despite resolution of the initial inflammatory exposure, demonstrating the persistence of the maladaptive impact induced by the inflammatory encounter. Thus, inflammation imprints the epigenome of ISCs in a manner that persists and affects their sensitivity to adapt to future stress or challenges.
Keywords: allogeneic hematopoietic stem cell transplantation; epigenetics; epithelial cell memory; graft-versus-host disease; intestinal stem cells; intestine organoids; metabolism; oxidative phosphorylation.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

