Identification and functional characterization of bidirectional gene pairs and their intergenic regions in cotton
- PMID: 39232709
- PMCID: PMC11373494
- DOI: 10.1186/s12870-024-05548-w
Identification and functional characterization of bidirectional gene pairs and their intergenic regions in cotton
Abstract
Background: In research to improve the quality of transgenic crops, it is often necessary to introduce multiple functionally related genes into recipient plants simultaneously to improve crop genetic traits effectively. Compared with unidirectional promoters, bidirectional promoters simultaneously regulate the expression of multiple genes and improve the efficiency of biotechnology. Therefore, in this study, bidirectional gene pairs were systematically analyzed in Gossypium hirsutum TM-1, and the structure, function and evolutionary relationships of the bidirectional genes were analyzed. The endogenous bidirectional promoters of cotton were mined, and their specific regulatory elements and biological functions were explored to provide useful promoter resources and a theoretical basis for cultivating new cotton germplasms with excellent fiber quality.
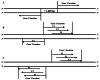
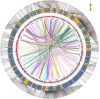
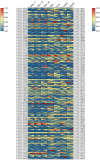
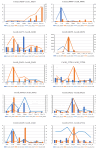
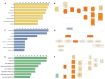
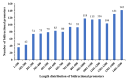
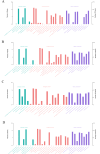
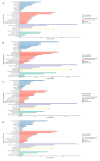
Results: Using an improved search model, a total of 1,383 bidirectional transcript pairs were identified in the Gossypium hirsutum TM-1 genome, and their gene structure and functional annotations were systematically analyzed. Thirty bidirectional intergenic sequences were randomly screened for promoter activity analysis via a transient expression system, and 25 intergenic sequences were found to have bidirectional promoter activity. Comparative analysis of the bidirectional gene profiles of the four cotton subspecies revealed that these subspecies presented abundant bidirectional gene pairs with high homology and that the bidirectional genes in the cotton subspecies were more similar in terms of their molecular functions, cellular components and biological processes. In addition, parallel analysis of bidirectional genes in dicotyledons and monocotyledons revealed that abundant bidirectional gene pairs exist in different species. Although the total number of orthologous bidirectional genes was similar, there was a significant difference in the number of orthologous bidirectional gene pairs between dicotyledons and monocotyledons. This evolutionary analysis of the function and structure of homologous bidirectional gene pairs in different varieties and different subspecies of the same species revealed potential pathways by which these gene pairs originated, which may be necessary for the evolution of a new species.
Conclusion: In this study, many bidirectional gene pairs in Gossypium hirsutum TM-1 were identified using computer programming, and systematic analysis was conducted to explore their functions and evolutionary relationships. In addition, the promoter activity of the bidirectional intergenic sequences was verified. The combination of computer programming screening, experimental validation and other methods is expected to provide preferred bidirectional promoters for transgenic breeding work via multigene cotransformation methods, and this information is valuable for genetic engineering research and applications.
Keywords: Gossypium hirsutum; Bidirectional gene pairs; Bidirectional promoter; Gene function; Genome-wide.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Structural and Functional Analysis of a Bidirectional Promoter from Gossypium hirsutum in Arabidopsis.Int J Mol Sci. 2018 Oct 23;19(11):3291. doi: 10.3390/ijms19113291. Int J Mol Sci. 2018. PMID: 30360512 Free PMC article.
-
Comparative transcriptome analysis of fiber and nonfiber tissues to identify the genes preferentially expressed in fiber development in Gossypium hirsutum.Sci Rep. 2021 Nov 24;11(1):22833. doi: 10.1038/s41598-021-01829-8. Sci Rep. 2021. PMID: 34819523 Free PMC article.
-
Genome-wide analysis and characterization of F-box gene family in Gossypium hirsutum L.BMC Genomics. 2019 Dec 19;20(1):993. doi: 10.1186/s12864-019-6280-2. BMC Genomics. 2019. PMID: 31856713 Free PMC article.
-
Identification and functional characterization of bidirectional gene pairs and their intergenic regions in maize.BMC Genomics. 2014 May 5;15(1):338. doi: 10.1186/1471-2164-15-338. BMC Genomics. 2014. PMID: 24886269 Free PMC article.
-
Genome-wide analyses of member identification, expression pattern, and protein-protein interaction of EPF/EPFL gene family in Gossypium.BMC Plant Biol. 2024 Jun 14;24(1):554. doi: 10.1186/s12870-024-05262-7. BMC Plant Biol. 2024. PMID: 38877405 Free PMC article.
Cited by
-
Through the lens of bioenergy crops: advances, bottlenecks, and promises of plant engineering.Plant J. 2025 Jul;123(2):e70294. doi: 10.1111/tpj.70294. Plant J. 2025. PMID: 40674648 Free PMC article. Review.
-
Unraveling the regulatory dynamics of bidirectional promoters for modulating gene co-expression and metabolic flux in Saccharomyces cerevisiae.Nucleic Acids Res. 2025 Jun 6;53(11):gkaf511. doi: 10.1093/nar/gkaf511. Nucleic Acids Res. 2025. PMID: 40503683 Free PMC article.
-
Plant engineering: advances, bottlenecks, and promise.Plant J. 2025 Apr;122(2):e70117. doi: 10.1111/tpj.70117. Plant J. 2025. PMID: 40275776
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

