A 3R-MYB transcription factor is involved in Methyl Jasmonate-Induced disease resistance in Agaricus bisporus and has implications for disease resistance in Arabidopsis
- PMID: 39233001
- PMCID: PMC12225956
- DOI: 10.1016/j.jare.2024.08.037
A 3R-MYB transcription factor is involved in Methyl Jasmonate-Induced disease resistance in Agaricus bisporus and has implications for disease resistance in Arabidopsis
Abstract
Introduction: Methyl jasmonate (MeJA) and MYB transcription factors (TFs) play important roles in pathogen resistance in several plants, but MYB TFs in conjunction with MeJA-induced defense against Pseudomonas tolaasii in edible mushrooms remain unknown.
Objectives: To investigate the role of a novel 3R-MYB transcription factor (AbMYB11) in MeJA-induced disease resistance of Agaricus bisporus and in the resistance of transgenic Arabidopsis to P. tolaasii.
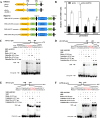
Methods: Mushrooms were treated with MeJA alone or in combination with phenylpropanoid pathway inhibitors, and the effects of the treatments on the disease-related and physiological indicators of the mushrooms were determined to assess the role of MeJA in inducing resistance and the importance of the phenylpropanoid pathway involved. Subcellular localization, gene expression analysis, dual-luciferase reporter assay, electrophoretic mobility shift assay, and transgenic Arabidopsis experiments were performed to elucidate the molecular mechanism of AbMYB11 in regulating disease resistance.
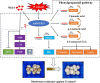
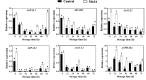
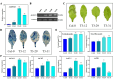
Results: MeJA application greatly improved mushroom resistance to P. tolaasii infection, and suppression of the phenylpropanoid pathway significantly weakened this effect. MeJA treatment stimulated the accumulation of phenylpropanoid metabolites, which was accompanied by increased the activities of biosynthetic enzymes and the expression of phenylpropanoid pathway-related genes (AbPAL1, Ab4CL1, AbC4H1) and an AbPR-like gene, further confirming the critical role of the phenylpropanoid pathway in MeJA-induced responses to P. tolaasii. Importantly, AbMYB11, localized in the nucleus, was rapidly induced by MeJA treatment under P. tolaasii infection; it transcriptionally activated the phenylpropanoid pathway-related and AbPR-like genes, and AbMYB11 overexpression in Arabidopsis significantly increased the transcription of phenylpropanoid-related genes, the accumulation of total phenolics and flavonoids, and improved resistance to P. tolaasii.
Conclusion: This study clarified the pivotal role of AbMYB11 as a regulator in disease resistance by modulating the phenylpropanoid pathway, providing a novel idea for the breeding of highly disease-resistant edible mushrooms and plants.
Keywords: Brown spot disease; Button mushroom; Methyl jasmonate; Phenylpropanoid metabolism; Pseudomonas tolaasii.
Copyright © 2024. Published by Elsevier B.V.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



References
-
- Marçal S., Sousa A.S., Taofiq O., Antunes F., Morais A.M.M.B., Freitas A.C., et al. Impact of postharvest preservation methods on nutritional value and bioactive properties of mushrooms. Trends Food Sci Technol. 2021;110:418–431. doi: 10.1016/j.tifs.2021.02.007. - DOI
-
- Li J., Wei Q., Huang L.X., Fang T., Chen B.Z., Jiang Y.J. Mathematical modeling Pseudomonas spp. growth and microflora composition variation in Agaricus bisporus fruiting bodies during chilled storage. Postharvest Biol Technol. 2020;163 doi: 10.1016/j.postharvbio.2020.111144. - DOI
-
- Li X., Ren Y.Y., Jing J., Jiang Y.R., Yang Q.L., Luo S.J., et al. The inhibitory mechanism of methyl jasmonate on Aspergillus flavus growth and aflatoxin biosynthesis and two novel transcription factors are involved in this action. Food Res Int. 2021;140 doi: 10.1016/j.foodres.2020.110051. - DOI - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

