Dual-species proteomics and targeted intervention of animal-pathogen interactions
- PMID: 39233003
- PMCID: PMC12225907
- DOI: 10.1016/j.jare.2024.08.038
Dual-species proteomics and targeted intervention of animal-pathogen interactions
Abstract
Introduction: Host-microbe interactions are important to human health and ecosystems globally, so elucidating the complex host-microbe interactions and associated protein expressions drives the need to develop sensitive and accurate biochemical techniques. Current proteomics techniques reveal information from the point of view of either the host or microbe, but do not provide data on the corresponding partner. Moreover, it remains challenging to simultaneously study host-microbe proteomes that reflect the direct competition between host and microbe. This raises the need to develop a dual-species proteomics method for host-microbe interactions.
Objectives: We aim to establish a forward + reverse Stable Isotope Labeling with Amino acids in Cell culture (SILAC) proteomics approach to simultaneously label and quantify newly-expressed proteins of host and microbe without physical isolation, for investigating mechanisms in direct host-microbe interactions.
Methods: Using Caenorhabditis elegans-Pseudomonas aeruginosa infection model as proof-of-concept, we employed SILAC proteomics and molecular pathway analysis to characterize the differentially-expressed microbial and host proteins. We then used molecular docking and chemical characterization to identify chemical inhibitors that intercept host-microbe interactions and eliminate microbial infection.
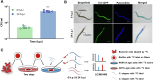
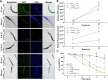
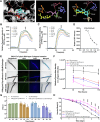
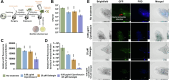
Results: Based on our proteomics results, we studied the iron competition between pathogen iron scavenger and host iron uptake protein, where P. aeruginosa upregulated pyoverdine synthesis protein (PvdA) (fold-change of 5.2313) and secreted pyoverdine, and C. elegans expressed ferritin (FTN-2) (fold-change of 3.4057). Targeted intervention of iron competition was achieved using Galangin, a ginger-derived phytochemical that inhibited pyoverdine production and biofilm formation in P. aeruginosa. The Galangin-ciprofloxacin combinatorial therapy could eliminate P. aeruginosa biofilms in a fish wound infection model, and enabled animal survival.
Conclusion: Our work provides a novel SILAC-based proteomics method that can simultaneously evaluate host and microbe proteomes, with future applications in higher host organisms and other microbial species. It also provides insights into the mechanisms dictating host-microbe interactions, offering novel strategies for anti-infective therapy.
Keywords: Caenorhabditis elegans; Host-microbe interactions; Iron competition; Pseudomonas aeruginosa; SILAC proteomics.
Copyright © 2024. Published by Elsevier B.V.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



Similar articles
-
Antibiotic strategies for eradicating Pseudomonas aeruginosa in people with cystic fibrosis.Cochrane Database Syst Rev. 2017 Apr 25;4(4):CD004197. doi: 10.1002/14651858.CD004197.pub5. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2023 Jun 2;6:CD004197. doi: 10.1002/14651858.CD004197.pub6. PMID: 28440853 Free PMC article. Updated.
-
Standard versus biofilm antimicrobial susceptibility testing to guide antibiotic therapy in cystic fibrosis.Cochrane Database Syst Rev. 2017 Oct 5;10(10):CD009528. doi: 10.1002/14651858.CD009528.pub4. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jun 10;6:CD009528. doi: 10.1002/14651858.CD009528.pub5. PMID: 28981972 Free PMC article. Updated.
-
Pseudomonas aeruginosa modulates both Caenorhabditis elegans attraction and pathogenesis by regulating nitrogen assimilation.Nat Commun. 2024 Sep 10;15(1):7927. doi: 10.1038/s41467-024-52227-3. Nat Commun. 2024. PMID: 39256376 Free PMC article.
-
Immunogenicity and seroefficacy of pneumococcal conjugate vaccines: a systematic review and network meta-analysis.Health Technol Assess. 2024 Jul;28(34):1-109. doi: 10.3310/YWHA3079. Health Technol Assess. 2024. PMID: 39046101 Free PMC article.
-
Salmonella exploits host- and bacterial-derived β-alanine for replication inside host macrophages.Elife. 2025 Jun 19;13:RP103714. doi: 10.7554/eLife.103714. Elife. 2025. PMID: 40536105 Free PMC article.
Cited by
-
Antibiotic-Resistant Pseudomonas aeruginosa: Current Challenges and Emerging Alternative Therapies.Microorganisms. 2025 Apr 16;13(4):913. doi: 10.3390/microorganisms13040913. Microorganisms. 2025. PMID: 40284749 Free PMC article. Review.
-
Bacterivorous nematodes decipher microbial iron siderophores as prey cue in predator-prey interactions.Proc Natl Acad Sci U S A. 2024 Jan 16;121(3):e2314077121. doi: 10.1073/pnas.2314077121. Epub 2024 Jan 8. Proc Natl Acad Sci U S A. 2024. PMID: 38190542 Free PMC article.
-
Lactobacilli-host interactions inhibit Staphylococcus aureus and Escherichia coli-induced cell death and invasion in a cellular model of infection.Front Microbiol. 2024 Dec 18;15:1501119. doi: 10.3389/fmicb.2024.1501119. eCollection 2024. Front Microbiol. 2024. PMID: 39744399 Free PMC article.
References
-
- Bottek J., Soun C., Lill J.K., Dixit A., Thiebes S., Beerlage A.-L., et al. Spatial proteomics revealed a CX3CL1-dependent crosstalk between the urothelium and relocated macrophages through IL-6 during an acute bacterial infection in the urinary bladder. Mucosal Immunol. 2020;13(4):702–714. - PMC - PubMed
-
- Kamaladevi A., Marudhupandiyan S., Balamurugan K. Model system based proteomics to understand the host response during bacterial infections. Mol Biosyst. 2017;13(12):2489–2497. - PubMed
-
- Munday D.C., Surtees R., Emmott E., Dove B.K., Digard P., Barr J.N., et al. Using SILAC and quantitative proteomics to investigate the interactions between viral and host proteomes. Proteomics. 2012;12(4–5):666–672. - PubMed
-
- Shi L., Adkins J.N., Coleman J.R., Schepmoes A.A., Dohnkova A., Mottaz H.M., et al. Proteomic analysis of Salmonella enterica serovar typhimurium isolated from RAW 264.7 macrophages: identification of a novel protein that contributes to the replication of serovar typhimurium inside macrophages. J Biol Chem. 2006;281(39):29131–29140. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

