Exploring and exploiting the rice phytobiome to tackle climate change challenges
- PMID: 39233440
- PMCID: PMC11671768
- DOI: 10.1016/j.xplc.2024.101078
Exploring and exploiting the rice phytobiome to tackle climate change challenges
Abstract
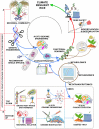
The future of agriculture is uncertain under the current climate change scenario. Climate change directly and indirectly affects the biotic and abiotic elements that control agroecosystems, jeopardizing the safety of the world's food supply. A new area that focuses on characterizing the phytobiome is emerging. The phytobiome comprises plants and their immediate surroundings, involving numerous interdependent microscopic and macroscopic organisms that affect the health and productivity of plants. Phytobiome studies primarily focus on the microbial communities associated with plants, which are referred to as the plant microbiome. The development of high-throughput sequencing technologies over the past 10 years has dramatically advanced our understanding of the structure, functionality, and dynamics of the phytobiome; however, comprehensive methods for using this knowledge are lacking, particularly for major crops such as rice. Considering the impact of rice production on world food security, gaining fresh perspectives on the interdependent and interrelated components of the rice phytobiome could enhance rice production and crop health, sustain rice ecosystem function, and combat the effects of climate change. Our review re-conceptualizes the complex dynamics of the microscopic and macroscopic components in the rice phytobiome as influenced by human interventions and changing environmental conditions driven by climate change. We also discuss interdisciplinary and systematic approaches to decipher and reprogram the sophisticated interactions in the rice phytobiome using novel strategies and cutting-edge technology. Merging the gigantic datasets and complex information on the rice phytobiome and their application in the context of regenerative agriculture could lead to sustainable rice farming practices that are resilient to the impacts of climate change.
Keywords: artificial intelligence; climate change; microbial ecology; rhizosphere engineering; rice microbiome; rice phytobiome.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures



References
-
- Aitchison J. The statistical analysis of compositional data. J. Roy. Stat. Soc. B. 1982;44:139–160. doi: 10.1111/j.2517-6161.1982.tb01195.x. - DOI
-
- Akatsuka T., Kodama O., Sekido H., Kono Y., Takeuchi S. Novel phytoalexins (oryzalexins A, B and C) isolated from rice blast leaves infected with Pyricularia oryzae. Part I: Isolation, characterization and biological activities of oryzalexins. Agricultural and biological chemistry. 1985;49:1689–1694. doi: 10.1080/00021369.1985.10866951. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

