TR-107, an Agonist of Caseinolytic Peptidase Proteolytic Subunit, Disrupts Mitochondrial Metabolism and Inhibits the Growth of Human Colorectal Cancer Cells
- PMID: 39233476
- PMCID: PMC11614700
- DOI: 10.1158/1535-7163.MCT-24-0170
TR-107, an Agonist of Caseinolytic Peptidase Proteolytic Subunit, Disrupts Mitochondrial Metabolism and Inhibits the Growth of Human Colorectal Cancer Cells
Abstract
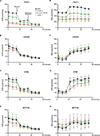
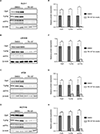
Oxidative phosphorylation is an essential metabolic process for cancer proliferation and therapy resistance. The ClpXP complex maintains mitochondrial proteostasis by degrading misfolded proteins. Madera Therapeutics has developed a class of highly potent and selective small-molecule activators (TR compounds) of the ClpXP component caseinolytic peptidase proteolytic subunit (ClpP). This approach to cancer therapy eliminates substrate recognition and activates nonspecific protease function within mitochondria, which has shown encouraging preclinical efficacy in multiple malignancies. The class-leading compound TR-107 has demonstrated significantly improved potency in ClpP affinity and activation and enhanced pharmacokinetic properties over the multitargeting clinical agent ONC201. In this study, we investigate the in vitro efficacy of TR-107 against human colorectal cancer cells. TR-107 inhibited colorectal cancer cell proliferation in a dose- and time-dependent manner and induced cell cycle arrest at low nanomolar concentrations. Mechanistically, TR-107 downregulated the expression of proteins involved in the mitochondrial unfolded protein response and mitochondrial DNA transcription and translation. TR-107 attenuated oxygen consumption rate and glycolytic compensation, confirming inactivation of oxidative phosphorylation and a reduction in total cellular respiration. Multiomics analysis of treated cells indicated a downregulation of respiratory chain complex subunits and an upregulation of mitophagy and ferroptosis pathways. Further evaluation of ferroptosis revealed a depletion of antioxidant and iron toxicity defenses that could potentiate sensitivity to combinatory chemotherapeutics. Together, this study provides evidence and insight into the subcellular mechanisms employed by colorectal cancer cells in response to potent ClpP agonism. Our findings demonstrate a productive approach to disrupting mitochondrial metabolism, supporting the translational potential of TR-107.
©2024 American Association for Cancer Research.
Conflict of interest statement
Conflict of interest disclosure:
EJI has an ownership interest in Madera Therapeutics, LLC. All other authors declare no potential conflicts of interest.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

