Identification of PI3K-AKT Pathway-Related Genes and Construction of Prognostic Prediction Model for ccRCC
- PMID: 39233640
- PMCID: PMC11375326
- DOI: 10.1002/cnr2.70010
Identification of PI3K-AKT Pathway-Related Genes and Construction of Prognostic Prediction Model for ccRCC
Abstract
Background: Clear cell renal cell carcinoma (ccRCC), the predominate histological type of renal cell carcinoma (RCC), has been extensively studied, with poor prognosis as the stage increases. Research findings consistently indicated that the PI3K-Akt pathway is commonly dysregulated across various cancer types, including ccRCC. Targeting the PI3K-Akt pathway held promise as a potential therapeutic approach for treating ccRCC. Development and validation of PI3K-Akt pathway-related genes related biomarkers can enhance healthcare management of patients with ccRCC.
Purpose: This study aimed to identify the key genes in the PI3K-Akt pathway associated with the diagnosis and prognosis of CCRCC using data mining from the Cancer Genome Atlas (TCGA) and Gene Expression Synthesis (GEO) datasets.
Methods: The purpose of this study is to use bioinformatics methods to screen data sets and clinicopathological characteristics associated with ccRCC patients. The exhibited significantly differential expressed genes (DEGs) associated with the PI3K-Akt pathway were examined by KEGG. In addition, Kaplan-Meier (KM) analysis used to estimate the survival function of the differential genes by using the UALCAN database and graphPad Prism 9.0. And exploring the association between the expression levels of the selected genes and the survival status and time of patients with ccRCC based on SPSS22.0. Finally, a multigene prognostic model was constructed to assess the prognostic risk of ccRCC patients.
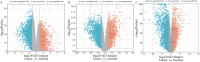
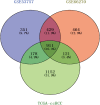
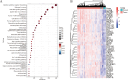
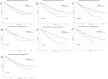
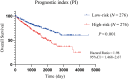
Results: A total of 911 genes with common highly expressed were selected based on the GEO and TCGA databases. According to the KEGG pathway analysis, there were 42 genes enriched in PI3K-Akt signalling pathway. And seven of highly expressed genes were linked to a poor prognosis in ccRCC. And a multigene prognostic model was established based on IL2RG, EFNA3, and MTCP1 synergistic expression might be utilized to predict the survival of ccRCC patients.
Conclusions: Three PI3K-Akt pathway-related genes may be helpful to identify the prognosis and molecular characteristics of ccRCC patients and to improve therapeutic regimens, and these risk characteristics might be further applied in the clinic.
Keywords: EFNA3; IL2RG; MTCP1; bioinformatics; clear cell renal cell carcinoma; prognostic model; the PI3K‐Akt signaling pathway.
© 2024 The Author(s). Cancer Reports published by Wiley Periodicals LLC.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


References
-
- Sung H., Ferlay J., Siegel R. L., et al., “Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries,” CA: A Cancer Journal for Clinicians 71, no. 3 (2021): 209–249. - PubMed
-
- Siegel R. L., Miller K. D., Fuchs H. E., and Jemal A., “Cancer Statistics, 2021,” CA: A Cancer Journal for Clinicians 71, no. 1 (2021): 7–33. - PubMed
-
- Gulati S. and Vaishampayan U., “Current State of Systemic Therapies for Advanced Renal Cell Carcinoma,” Current Oncology Reports 22, no. 3 (2020): 26. - PubMed
-
- Fisher R., Gore M., and Larkin J., “Current and Future Systemic Treatments for Renal Cell Carcinoma,” Seminars in Cancer Biology 23, no. 1 (2013): 38–45. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

