Preservation of cfRNA in cytological supernatants for cfDNA & cfRNA double detection in non-small cell lung cancer patients
- PMID: 39233657
- PMCID: PMC11375324
- DOI: 10.1002/cam4.70197
Preservation of cfRNA in cytological supernatants for cfDNA & cfRNA double detection in non-small cell lung cancer patients
Abstract
Backgroud: Supernatants from various cytological samples, including body cavity effusion, sputum, bronchoalveolar lavage fluid (BALF), and needle aspiration, have been validated for detecting genetic alterations using cell-free DNA (cfDNA) in patients with non-small cell lung cancer (NSCLC). However, the sensitivity of fusion variations detection remains challenging. The protection of cell-free RNA (cfRNA) is critical for resolving the issue.
Methods: A protective solution (PS) was applied for preserving cfRNA in cytological supernatant (CS), and the quality of protected cfRNA was assessed by cycle threshold (CT) values from reverse transcription quantitative polymerase chain reaction (RT-qPCR). Furthermore, we collected an additional set of malignant cytological and matched tumor samples from 84 NSCLC patients, cfDNA & cfRNA extraction and double detection for driver gene mutations was validated using the multi-gene mutations detection by RT-qPCR.
Results: Under the optimal protection system, 91.0% (101/111) of cfRNA were protected effectively. Among the 84 NSCLC patient samples, seven cytological samples failed the tests. In comparison with tumor samples, the overall sensitivity and specificity of detecting driver genes of supernatant cfDNA and cfRNA were 93.8% (74/77) and 100% (77/77), respectively. Notably, when focusing exclusively on patients with fusion gene changes, both sensitivity and specificity reached 100% (11/11) for EML4-ALK, ROS1, RET fusions, and MET ex14 skipping.
Conclusion: These findings suggest that cfDNA & cfRNA extraction and double detection strategy recommended in this study improve the accuracy of driver genes mutations test, especially for RNA-based assay.
Keywords: NSCLC; cell‐free RNA; driver gene; fusion; liquid biopsy.
© 2024 The Author(s). Cancer Medicine published by John Wiley & Sons Ltd.
Figures
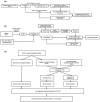

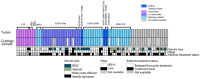
References
-
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394‐424. - PubMed
-
- Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131(6):1190‐1203. - PubMed
-
- Li YS, Jiang BY, Yang JJ, et al. Unique genetic profiles from cerebrospinal fluid cell‐free DNA in leptomeningeal metastases of EGFR‐mutant non‐small‐cell lung cancer: a new medium of liquid biopsy. Ann Oncol. 2018;29(4):945‐952. - PubMed
-
- Wang Z, Zhang L, Li L, et al. Sputum cell‐free DNA: valued surrogate sample for detection of EGFR mutation in patients with advanced lung adenocarcinoma. J Mol Diagn. 2020;22(7):934‐942. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

