Single-cell RNA sequencing of pediatric Hodgkin lymphoma to study the inhibition of T cell subtypes
- PMID: 39233904
- PMCID: PMC11369206
- DOI: 10.1002/hem3.149
Single-cell RNA sequencing of pediatric Hodgkin lymphoma to study the inhibition of T cell subtypes
Erratum in
-
Correction to "Single-cell RNA sequencing of pediatric Hodgkin lymphoma to study the inhibition of T cell subtypes".Hemasphere. 2024 Oct 7;8(10):e70023. doi: 10.1002/hem3.70023. eCollection 2024 Oct. Hemasphere. 2024. PMID: 39380840 Free PMC article.
Abstract
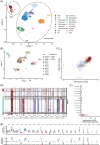
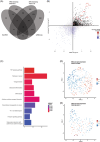
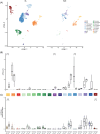
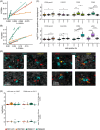
Pediatric classic Hodgkin lymphoma (cHL) patients have a high survival rate but suffer from severe long-term side effects induced by chemo- and radiotherapy. cHL tumors are characterized by the low fraction (0.1%-10%) of malignant Hodgkin and Reed-Sternberg (HRS) cells in the tumor. The HRS cells depend on the surrounding immune cells for survival and growth. This dependence is leveraged by current treatments that target the PD-1/PD-L1 axis in cHL tumors. The development of more targeted therapies that are specific for the tumor and are therefore less toxic for healthy tissue compared with conventional chemotherapy could improve the quality of life of pediatric cHL survivors. Here, we applied single-cell RNA sequencing (scRNA-seq) on isolated HRS cells and the immune cells from the same cHL tumors. Besides TNFRSF8 (CD30), we identified other genes of cell surface proteins that are consistently overexpressed in HRS cells, such as NRXN3 and LRP8, which can potentially be used as alternative targets for antibody-drug conjugates or CAR T cells. Finally, we identified potential interactions by which HRS cells inhibit T cells, among which are the galectin-1/CD69 and HLA-II/LAG3 interactions. RNAscope was used to validate the enrichment of CD69 and LAG3 expression on T cells near HRS cells and indicated large variability of the interaction strength with the corresponding ligands between patients and between tumor tissue regions. In conclusion, this study identifies new potential therapeutic targets for cHL and highlights the importance of studying heterogeneity when identifying therapy targets, specifically those that target tumor-immune cell interactions.
© 2024 The Author(s). HemaSphere published by John Wiley & Sons Ltd on behalf of European Hematology Association.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
-
- Brice P, de Kerviler E, Friedberg JW. Classical Hodgkin lymphoma. Lancet. 2021;398:1518‐1527. - PubMed
-
- LaCasce AS. Treating Hodgkin lymphoma in the new millennium: relapsed and refractory disease. Hematol Oncol. 2019;37:87‐91. - PubMed
-
- Raut M, Singh G, Hiscock I, Sharma S, Pilkhwal N. A systematic literature review of the epidemiology, quality of life, and economic burden, including disease pathways and treatment patterns of relapsed/refractory classical Hodgkin lymphoma. Expert Rev Hematol. 2022;15:607‐617. - PubMed
-
- Ehrhardt MJ, Flerlage JE, Armenian SH, Castellino SM, Hodgson DC, Hudson MM. Integration of pediatric Hodgkin lymphoma treatment and late effects guidelines: seeing the forest beyond the trees. J Natl Compr Cancer Netw. 2021;19:755‐764. - PubMed
-
- Ansell SM. Hodgkin lymphoma: 2018 update on diagnosis, risk‐stratification, and management. Am J Hematol. 2018;93:704‐715. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
