A cellular disease model toward gene therapy of TGM1-dependent lamellar ichthyosis
- PMID: 39234443
- PMCID: PMC11372595
- DOI: 10.1016/j.omtm.2024.101311
A cellular disease model toward gene therapy of TGM1-dependent lamellar ichthyosis
Abstract
Lamellar ichthyosis (LI) is a chronic disease, mostly caused by mutations in the TGM1 gene, marked by impaired skin barrier formation. No definitive therapies are available, and current treatments aim at symptomatic relief. LI mouse models often fail to faithfully replicate the clinical and histopathological features of human skin conditions. To develop advanced therapeutic approaches, such as combined ex vivo cell and gene therapy, we established a human cellular model of LI by efficient CRISPR-Cas9-mediated gene ablation of the TGM1 gene in human primary clonogenic keratinocytes. Gene-edited cells showed complete absence of transglutaminase 1 (TG1) expression and recapitulated a hyperkeratotic phenotype with most of the molecular hallmarks of LI in vitro. Using a self-inactivating γ-retroviral (SINγ-RV) vector expressing transgenic TGM1 under the control of its own promoter, we tested an ex vivo gene therapy approach and validate the model of LI as a platform for pre-clinical evaluation studies. Gene-corrected TGM1-null keratinocytes displayed proper TG1 expression, enzymatic activity, and cornified envelope formation and, hence, restored proper epidermal architecture. Single-cell multiomics analysis demonstrated proviral integrations in holoclone-forming epidermal stem cells, which are crucial for epidermal regeneration. This study serves as a proof of concept for assessing the potential of this therapeutic approach in treating TGM1-dependent LI.
Keywords: 3D skin equivalent; CRISPR-Cas9; TGM1; gene therapy; lamellar ichthyosis; multiomics analysis; primary human keratinocytes; regenerative medicine.
© 2024 The Author(s).
Conflict of interest statement
M.D.L. is a consultant for J-TEC-Japan Tissue Engineering, Ltd.
Figures

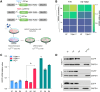





References
-
- Oji V., Tadini G., Akiyama M., Blanchet Bardon C., Bodemer C., Bourrat E., Coudiere P., DiGiovanna J.J., Elias P., Fischer J., et al. Revised nomenclature and classification of inherited ichthyoses: results of the First Ichthyosis Consensus Conference in Soreze 2009. J. Am. Acad. Dermatol. 2010;63:607–641. - PubMed
-
- Abernethy J.L., Hill R.L., Goldsmith L.A. epsilon-(gamma-Glutamyl) lysine cross-links in human stratum corneum. J. Biol. Chem. 1977;252:1837–1839. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

