Spatial Transcriptomic Approach to Understanding Coronary Atherosclerotic Plaque Stability
- PMID: 39234691
- PMCID: PMC11499036
- DOI: 10.1161/ATVBAHA.123.320330
Spatial Transcriptomic Approach to Understanding Coronary Atherosclerotic Plaque Stability
Abstract
Background: Coronary atherosclerotic plaques susceptible to acute coronary syndrome have traditionally been characterized by their surrounding cellular architecture. However, with the advent of intravascular imaging, novel mechanisms of coronary thrombosis have emerged, challenging our contemporary understanding of acute coronary syndrome. These intriguing findings underscore the necessity for a precise molecular definition of plaque stability. Considering this, our study aimed to investigate the vascular microenvironment in patients with stable and unstable plaques using spatial transcriptomics.
Methods: Autopsy-derived coronary arteries were preserved and categorized by plaque stability (n=5 patients per group). We utilized the GeoMx spatial profiling platform and Whole Transcriptome Atlas to link crucial histological morphology markers in coronary lesions with differential gene expression in specific regions of interest, thereby mapping the vascular transcriptome. This innovative approach allowed us to conduct cell morphological and spatially resolved transcriptional profiling of atherosclerotic plaques while preserving crucial intercellular signaling.
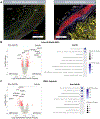
Results: We observed intriguing spatial and cell-specific transcriptional patterns in stable and unstable atherosclerotic plaques, showcasing regional variations within the intima and media. These regions exhibited differential expression of proinflammatory molecules (eg, IFN-γ [interferon-γ], MHC [major histocompatibility complex] class II, proinflammatory cytokines) and prothrombotic signaling pathways. By using lineage tracing through spatial deconvolution of intimal CD68+ (cluster of differentiation 68) cells, we characterized unique, intraplaque subpopulations originating from endothelial, smooth muscle, and myeloid lineages with distinct regional locations associated with plaque instability. In addition, unique transcriptional signatures were observed in vascular smooth muscle and CD68+ cells among plaques exhibiting coronary calcification.
Conclusions: Our study illuminates distinct cell-specific and regional transcriptional alterations present in unstable plaques. Furthermore, we characterize spatially resolved, in situ evidence supporting cellular transdifferentiation and intraplaque plasticity as significant contributors to plaque instability in human coronary atherosclerosis. Our results provide a powerful resource for the identification of novel mediators of acute coronary syndrome, opening new avenues for preventative and therapeutic treatments.
Keywords: acute coronary syndrome; atherosclerosis; coronary thrombosis; macrophages; muscle, smooth, vascular.
Conflict of interest statement
None.
Figures




Comment in
-
Spatial Transcriptomics: A New Frontier in Atherosclerosis Research?Arterioscler Thromb Vasc Biol. 2024 Nov;44(11):2291-2293. doi: 10.1161/ATVBAHA.124.321652. Epub 2024 Oct 23. Arterioscler Thromb Vasc Biol. 2024. PMID: 39441914 No abstract available.
References
-
- Hagstrom E, Steg PG, Szarek M, Bhatt DL, Bittner VA, Danchin N, Diaz R, Goodman SG, Harrington RA, Jukema JW, et al. Apolipoprotein B, Residual Cardiovascular Risk After Acute Coronary Syndrome, and Effects of Alirocumab. Circulation. 2022;146:657–672. doi: 10.1161/CIRCULATIONAHA.121.057807 - DOI - PMC - PubMed
-
- Tong DC, Quinn S, Nasis A, Hiew C, Roberts-Thomson P, Adams H, Sriamareswaran R, Htun NM, Wilson W, Stub D, et al. Colchicine in Patients With Acute Coronary Syndrome: The Australian COPS Randomized Clinical Trial. Circulation. 2020;142:1890–1900. doi: 10.1161/CIRCULATIONAHA.120.050771 - DOI - PubMed
-
- Schwartz GG, Ballantyne CM, Barter PJ, Kallend D, Leiter LA, Leitersdorf E, McMurray JJV, Nicholls SJ, Olsson AG, Shah PK, et al. Association of Lipoprotein(a) With Risk of Recurrent Ischemic Events Following Acute Coronary Syndrome: Analysis of the dal-Outcomes Randomized Clinical Trial. JAMA Cardiol. 2018;3:164–168. doi: 10.1001/jamacardio.2017.3833 - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

