FGF12 Positively Regulates Keratinocyte Proliferation by Stabilizing MDM2 and Inhibiting p53 Activity in Psoriasis
- PMID: 39234815
- PMCID: PMC11497104
- DOI: 10.1002/advs.202400107
FGF12 Positively Regulates Keratinocyte Proliferation by Stabilizing MDM2 and Inhibiting p53 Activity in Psoriasis
Abstract
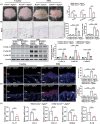
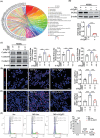
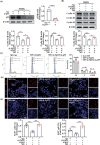
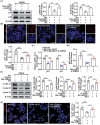
Psoriasis is a chronic skin disease characterized by abnormal proliferation and inflammation of epidermal keratinocytes. Fibroblast growth factor 12 (FGF12) is implicated in the regulation of diverse cellular signals; however, its precise mechanism in psoriasis requires further investigation. In this study, high expression of FGF12 is observed in the epidermis of skin lesion in psoriasis patients and imiquimod (IMQ)-induced psoriasis like-dermatitis. Moreover, specific loss of FGF12 in keratinocytes in IMQ-induced psoriasis model alleviates psoriasis-like symptoms and reduces proliferation. In vitro RNA sequencing demonstrates that knockdown of FGF12 effectively arrests the cell cycle, inhibits cell proliferation, and predominantly regulates the p53 signaling pathway. Mechanistically, FGF12 is selectively bound to the RING domain of MDM2, thus partially inhibiting the binding of β-Trcp to MDM2. This interaction inhibits β-Trcp-induced-K48 ubiquitination degradation of MDM2, thereby suppressing the activity of the p53 signaling pathway, which results in excessive cell proliferation. Last, the alleviatory effect of FGF12 deficiency on psoriasis progression is reversed by p53 knockdown. In summary, these findings provide valuable insights into the mechanisms by which FGF12 suppresses p53 signaling in keratinocytes, exacerbating the development of psoriasis. This positive regulatory loop highlights the potential of FGF12 as a therapeutic target to manage psoriasis.
Keywords: FGF12; MDMD2; p53; proliferation; psoriasis.
© 2024 The Author(s). Advanced Science published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Cordycepin Ameliorates Psoriasis-Like Skin Lesion by Regulating p53/MDM2 Feedback Loop.Mol Biotechnol. 2025 Jun;67(6):2481-2492. doi: 10.1007/s12033-024-01211-9. Epub 2024 Jun 24. Mol Biotechnol. 2025. PMID: 38914920
-
Sinomenine Suppressed Keratinocyte Proliferation and Imiquimod-Induced Psoriasis-Like Dermatitis by Regulating lncRNA XIST.Skin Pharmacol Physiol. 2022;35(6):328-342. doi: 10.1159/000526420. Epub 2022 Aug 12. Skin Pharmacol Physiol. 2022. PMID: 35960313
-
Switch from p53 to MDM2 as differentiating human keratinocytes lose their proliferative potential and increase in cellular size.Oncogene. 2000 Aug 3;19(33):3693-705. doi: 10.1038/sj.onc.1203695. Oncogene. 2000. PMID: 10949923
-
Lysophosphatidic Acid Mediates Imiquimod-Induced Psoriasis-like Symptoms by Promoting Keratinocyte Proliferation through LPAR1/ROCK2/PI3K/AKT Signaling Pathway.Int J Mol Sci. 2021 Oct 5;22(19):10777. doi: 10.3390/ijms221910777. Int J Mol Sci. 2021. PMID: 34639115 Free PMC article.
-
Mdm2 links genotoxic stress and metabolism to p53.Protein Cell. 2010 Dec;1(12):1063-72. doi: 10.1007/s13238-010-0140-9. Epub 2011 Jan 8. Protein Cell. 2010. PMID: 21213101 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
