BCAT1 is a NOTCH1 target and sustains the oncogenic function of NOTCH1
- PMID: 39234857
- PMCID: PMC11788623
- DOI: 10.3324/haematol.2024.285552
BCAT1 is a NOTCH1 target and sustains the oncogenic function of NOTCH1
Abstract
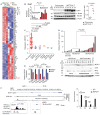
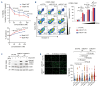
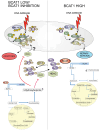
High levels of branched-chain amino acid (BCAA) transaminase 1 (BCAT1) have been associated with tumor aggressiveness and drug resistance in several cancer types. Nevertheless, the mechanistic role of BCAT1 in T-cell acute lymphoblastic leukemia (T-ALL) remains uncertain. We provide evidence that Bcat1 was over-expressed following NOTCH1-induced transformation of leukemic progenitors and that NOTCH1 directly controlled BCAT1 expression by binding to a BCAT1 promoter. Further, using a NOTCH1 gain-of-function retroviral model of T-ALL, mouse cells genetically deficient for Bcat1 showed defects in developing leukemia. In murine T-ALL cells, Bcat1 depletion or inhibition redirected leucine metabolism towards production of 3-hydroxy butyrate (3-HB), an endogenous histone deacetylase inhibitor. Consistently, BCAT1-depleted cells showed altered protein acetylation levels which correlated with a pronounced sensitivity to DNA damaging agents. In human NOTCH1-dependent leukemias, high expression levels of BCAT1 may predispose to worse prognosis. Therapeutically, BCAT1 inhibition specifically synergized with etoposide to eliminate tumors in patient-derived xenograft models suggesting that BCAT1 inhibitors may have a part to play in salvage protocols for refractory T-ALL.
Figures




References
-
- Pui CH, Robison LL, Look AT. Acute lymphoblastic leukaemia. Lancet. 2008;371(9617):1030-1043. - PubMed
-
- Belver L, Ferrando A. The genetics and mechanisms of T cell acute lymphoblastic leukaemia. Nature reviews Cancer. 2016;16(8):494-507. - PubMed
-
- Weng AP, Ferrando AA, Lee W, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306(5694):269-271. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

