Expression of Plasmodium major facilitator superfamily protein in transporters - Δ Candida identifies a drug transporter
- PMID: 39235058
- PMCID: PMC11485967
- DOI: 10.1080/17460913.2024.2389750
Expression of Plasmodium major facilitator superfamily protein in transporters - Δ Candida identifies a drug transporter
Abstract
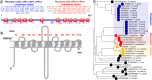
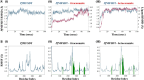
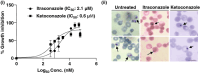
Aim: To assess the functional relevance of a putative Major Facilitator Superfamily protein (PF3D7_0210300; 'PfMFSDT') as a drug transporter, using Candida glabrata for orthologous protein expression.Methods: Complementary Determining Sequence encoding PfMFSDT was integrated into the genome of genetically engineered C. glabrata strain MSY8 via homologous recombination, followed by assessing its functional relevance as a drug transporter.Results & conclusion: The modified C. glabrata strain exhibited plasma membrane localization of PfMFSDT and characteristics of an Major Facilitator Superfamily transporter, conferring resistance to antifungals, ketoconazole and itraconazole. The nanomolar inhibitory effects of the drugs on the intra-erythrocytic growth of Plasmodium falciparum highlight their antimalarial properties. This study proposes PfMFSDT as a drug transporter, expanding the repertoire of the currently known antimalarial 'resistome'.
Keywords: Candida glabrata; Plasmodium falciparum; antimalarial; drug discovery; membrane transporter; orthologous system.
Plain language summary
[Box: see text].
Conflict of interest statement
NG acknowledges Department of Biotechnology, Government of India (Grant no. BT/PR46993/PBD/26/834/2023). PM is a recipient of Junior Research Fellowship from Department of Biotechnology, Government of India (Ref. no.: DBTHRDPMU/JRF/BET-21/I/2021-22/326). MK acknowledges Research Associate fellowship from Indian Council of Medical Research (ICMR-RA), Government of India (Myco/fell/3/2022-ECD-II). RJ acknowledges Research Associate I fellowship from Indo-DBT (Department of Biotechnology), Government of India and Swiss National Science foundation. AG acknowledges Laboratory Assistant fellowship from Indo-DBT (Department of Biotechnology), Government of India and Swiss National Science foundation. The authors have no other competing interests or relevant affiliations with any organization or entity with the subject matter or materials discussed in the manuscript apart from those disclosed.
Figures




Similar articles
-
The proton-dependent oligopeptide transporter Ptr_C serves as an efficient system for antifungal delivery into Candida auris.J Appl Microbiol. 2025 Jul 1;136(7):lxaf170. doi: 10.1093/jambio/lxaf170. J Appl Microbiol. 2025. PMID: 40622996
-
Inner-mitochondrial membrane protein PfMPV17 is linked to P. falciparum in vitro resistance to the indoloquinolizidine alkaloid alstonine.J Antimicrob Chemother. 2025 Jul 1;80(7):1869-1877. doi: 10.1093/jac/dkaf141. J Antimicrob Chemother. 2025. PMID: 40432501 Free PMC article.
-
Antifungal susceptibility profiles of Candida and non-albicans species isolated from pregnant women: implications for emerging antimicrobial resistance in maternal health.Microbiol Spectr. 2025 Jul;13(7):e0078725. doi: 10.1128/spectrum.00787-25. Epub 2025 Jun 2. Microbiol Spectr. 2025. PMID: 40454865 Free PMC article.
-
Primaquine for reducing Plasmodium falciparum transmission.Cochrane Database Syst Rev. 2012 Sep 12;(9):CD008152. doi: 10.1002/14651858.CD008152.pub2. Cochrane Database Syst Rev. 2012. Update in: Cochrane Database Syst Rev. 2014 Jun 30;(6):CD008152. doi: 10.1002/14651858.CD008152.pub3. PMID: 22972117 Updated.
-
Anti-folate drug resistance in Africa: meta-analysis of reported dihydrofolate reductase (dhfr) and dihydropteroate synthase (dhps) mutant genotype frequencies in African Plasmodium falciparum parasite populations.Malar J. 2010 Aug 30;9:247. doi: 10.1186/1475-2875-9-247. Malar J. 2010. PMID: 20799995 Free PMC article.
References
-
- World Health Organization (WHO) . World malaria report 2023 [Internet]. 2023. Available from: https://www.who.int/teams/global-malaria-programme/reports/world-malaria...
-
- McKee RW, Ormsbee RA, Anfinsen CB, et al. The chemistry and metabolism of normal and parasitized (P. knowlesi) monkey blood. J Exp Med. 1946;84:569–582. doi: 10.1084/jem.84.6.569 - DOI - PMC - PubMed
-
• These references discuss the metabolic pathways of the Plasmodium parasite and the potential for targeting these pathways for antimalarial development.
-
- Pfaller MA, Krogstad DJ, Parquette AR, et al. Plasmodium falciparum: stage-specific lactate production in synchronized cultures. Exp Parasitol. 1982;54:391–396. doi: 10.1016/0014-4894(82)90048-0 - DOI - PubMed
-
• These references discuss the metabolic pathways of the Plasmodium parasite and the potential for targeting these pathways for antimalarial development.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
