Use of 3M-052-AF with Alum adjuvant in HIV trimer vaccine induces human autologous neutralizing antibodies
- PMID: 39235529
- PMCID: PMC11380150
- DOI: 10.1084/jem.20240604
Use of 3M-052-AF with Alum adjuvant in HIV trimer vaccine induces human autologous neutralizing antibodies
Abstract
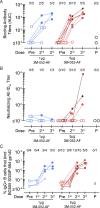
Stabilized trimers preserving the native-like HIV envelope structure may be key components of a preventive HIV vaccine regimen to induce broadly neutralizing antibodies (bnAbs). We evaluated trimeric BG505 SOSIP.664 gp140 formulated with a novel TLR7/8 signaling adjuvant, 3M-052-AF/Alum, for safety, adjuvant dose-finding, and immunogenicity in a first-in-healthy adult (n = 17), randomized, and placebo-controlled trial (HVTN 137A). The vaccine regimen appeared safe. Robust, trimer-specific antibody, and B cell and CD4+ T cell responses emerged after vaccination. Five vaccinees developed serum autologous tier 2 nAbs (ID50 titer, 1:28-1:8647) after two to three doses targeting C3/V5 and/or V1/V2/V3 Env regions by electron microscopy and mutated pseudovirus-based neutralization analyses. Trimer-specific, B cell-derived monoclonal antibody activities confirmed these results and showed weak heterologous neutralization in the strongest responder. Our findings demonstrate the clinical utility of the 3M-052-AF/Alum adjuvant and support further improvements of trimer-based Env immunogens to focus responses on multiple broad nAb epitopes.
© 2024 Hahn et al.
Conflict of interest statement
Disclosures: H. Janes reported grants from the National Institutes of Health during the conduct of the study and grants from the National Institutes of Health outside the submitted work. M. Tomai reported being an employee at 3M Company when this research was started and having received stock options while working at 3M. M. Tomai also received options from 3M while these studies were being performed. M. Tomai is now a contract employee for Solventum, which owns the vaccine adjuvant 3M-052 that was used in this study. L. Corey reported grants from NIAID/National Institutes of Health during the conduct of the study. S. Edupuganti reported grants from HIV Vaccine Trials Network during the conduct of the study. I. Frank reported grants from Gilead Sciences and Johnson and Johnson during the conduct of the study and personal fees from GlaxoSmithKline and Johnson and Johnson outside the submitted work. R.W. Sanders and J.P. Moore reported a patent to EP2765138A3/US20140212458 issued. A.B. Ward reported personal fees from Third Rock Ventures outside the submitted work. N. Rouphael reported grants from Emory University during the conduct of the study; and that Emory receives funds for N. Rouphael to conduct research from Sanofi, Lilly, Merck, Quidel, Immorna, Vaccine Company, and Pfizer. N. Rouphael served on selected advisory boards for Sanofi, Seqirus, Pfizer and Moderna and is a paid clinical trials safety consultant for ICON, CyanVac, Imunon, and EMMES. Fred Hutch Cancer Center receives funds for M.J. McElrath to conduct research from Janssen, Sanofi, Regeneron, and Moderna. No other disclosures were reported.
Figures






Update of
-
HIV BG505 SOSIP.664 trimer with 3M-052-AF/alum induces human autologous tier-2 neutralizing antibodies.medRxiv [Preprint]. 2024 May 9:2024.05.08.24306957. doi: 10.1101/2024.05.08.24306957. medRxiv. 2024. Update in: J Exp Med. 2024 Oct 7;221(10):e20240604. doi: 10.1084/jem.20240604. PMID: 38766048 Free PMC article. Updated. Preprint.
References
-
- Bianchi, M., Turner H.L., Nogal B., Cottrell C.A., Oyen D., Pauthner M., Bastidas R., Nedellec R., McCoy L.E., Wilson I.A., et al. 2018. Electron-microscopy-based epitope mapping defines specificities of polyclonal antibodies elicited during HIV-1 BG505 envelope trimer immunization. Immunity. 49:288–300.e8. 10.1016/j.immuni.2018.07.009 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

