Immune microenvironmental heterogeneity according to tumor DNA methylation phenotypes in microsatellite instability-high colorectal cancers
- PMID: 39235590
- PMCID: PMC11377388
- DOI: 10.1007/s00262-024-03805-3
Immune microenvironmental heterogeneity according to tumor DNA methylation phenotypes in microsatellite instability-high colorectal cancers
Abstract
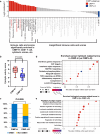
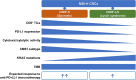
The detailed association between tumor DNA methylation, including CpG island methylation, and tumor immunity is poorly understood. CpG island methylator phenotype (CIMP) is observed typically in sporadic colorectal cancers (CRCs) with microsatellite instability-high (MSI-H). Here, we investigated the differential features of the tumor immune microenvironment according to CIMP status in MSI-H CRCs. CIMP-high (CIMP-H) or CIMP-low/negative (CIMP-L/0) status was determined using MethyLight assay in 133 MSI-H CRCs. All MSI-H CRCs were subjected to digital pathology-based quantification of CD3 + /CD8 + /CD4 + /FoxP3 + /CD68 + /CD204 + /CD177 + tumor-infiltrating immune cells using whole-slide immunohistochemistry. Programmed death-ligand 1 (PD-L1) immunohistochemistry was evaluated using the tumor proportion score (TPS) and combined positive score (CPS). Representative cases were analyzed using whole-exome and RNA-sequencing. In 133 MSI-H CRCs, significantly higher densities of CD8 + tumor-infiltrating lymphocytes (TILs) were observed in CIMP-H tumors compared with CIMP-L/0 tumors. PD-L1 TPS and CPS in CIMP-H tumors were higher than in CIMP-L/0 tumors. Next-generation sequencing revealed that, compared with CIMP-L/0 tumors, CIMP-H tumors had higher fractions of CD8 + T cells/cytotoxic lymphocytes, higher cytolytic activity scores, and activated immune-mediated cell killing pathways. In contrast to CIMP-L/0 tumors, most CIMP-H tumors were identified as consensus molecular subtype 1, an immunogenic transcriptomic subtype of CRC. However, there were no differences in tumor mutational burden (TMB) between CIMP-H and CIMP-L/0 tumors in MSI-H CRCs. In conclusion, CIMP-H is associated with abundant cytotoxic CD8 + TILs and PD-L1 overexpression independent of TMB in MSI-H CRCs, suggesting that CIMP-H tumors represent a typical immune-hot subtype and are optimal candidates for immunotherapy in MSI-H tumors.
Keywords: Colorectal carcinoma; CpG Island methylation; DNA methylation; Microsatellite instability; Mismatch repair; Tumor immunology.
© 2024. The Author(s).
Conflict of interest statement
The authors have no relevant financial or non-financial interests to disclose.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

