Escherichia coli ST117: exploring the zoonotic hypothesis
- PMID: 39235965
- PMCID: PMC11448156
- DOI: 10.1128/spectrum.00466-24
Escherichia coli ST117: exploring the zoonotic hypothesis
Abstract
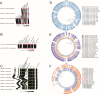
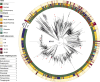
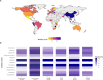
Extraintestinal pathogenic Escherichia coli (ExPEC) can lead to severe infections, with additional risks of increasing antimicrobial resistance rates. Genotypic similarities between ExPEC and avian pathogenic E. coli (APEC) support a possible role for a poultry meat reservoir in human disease. Some genomic studies have been done on the ST117 lineage which contaminates poultry meat, carries multidrug resistance, can be found in the human intestinal microbiota, and causes human extraintestinal disease. This study analyzed the genomes of 61 E. coli from Brazilian poultry outbreaks focusing on ST117, to further define its possible zoonotic characteristics by genotypic and phylogenomic analyses, along with 1,699 worldwide ST117 isolates originating from human, animal, and environment sources. A predominance of ST117 was detected in the Brazilian isolates (n = 20/61) frequently carrying resistance to critical antibiotics (>86%) linked to IncFII, IncI1, or IncX4 replicons. High similarities were found between IncX4 from Brazilian outbreaks and those from E. coli recovered from imported Brazilian poultry meat and human clinical cases. The ST117 phylogeny showed non-specificity according to host and continent and an AMR index score indicated the highest resistance in Asia and South America, with the latter statistically more resistant and overrepresented with resistance to extended-spectrum beta-lactamases (ESBL). Most ST117 human isolates were predicted to have a poultry origin (93%, 138/148). In conclusion, poultry is a likely source for zoonotic ExPEC strains, particularly the ST117 lineage which can also serve as a reservoir for resistance determinants against critical antibiotics encoded on highly transmissible plasmids.
Importance: Certain extraintestinal pathogenic Escherichia coli (ExPEC) are particularly important as they affect humans and animals. Lineages, such as ST117, are predominant in poultry and frequent carriers of antibiotic resistance, presenting a risk to humans handling or ingesting poultry products. We analyzed ExPEC isolates causing outbreaks in Brazilian poultry, focusing on the ST117 as the most detected lineage. Genomic comparisons with international isolates from humans and animals were performed describing the potential zoonotic profile. The Brazilian ST117 isolates carried resistance determinants against critical antibiotics, mainly on plasmids, in some cases identical to those carried by international isolates. South American ST117 isolates from all sources generally exhibit more resistance, including to critical antibiotics, and worldwide, the vast majority of human isolates belonging to this lineage have a predicted poultry origin. As the world's largest poultry exporter, Brazil has an important role in developing strategies to prevent the dissemination of multidrug-resistant zoonotic ExPEC strains.
Keywords: Escherichia coli; ExPEC; FZEC; ST117; poultry.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Kidsley AK, O’Dea M, Ebrahimie E, Mohammadi-Dehcheshmeh M, Saputra S, Jordan D, Johnson JR, Gordon D, Turni C, Djordjevic SP, Abraham S, Trott DJ. 2020. Genomic analysis of fluoroquinolone-susceptible phylogenetic group B2 extraintestinal pathogenic Escherichia coli causing infections in cats. Vet Microbiol 245:108685. doi: 10.1016/j.vetmic.2020.108685 - DOI - PubMed
-
- Kidsley AK, O’Dea M, Saputra S, Jordan D, Johnson JR, Gordon DM, Turni C, Djordjevic SP, Abraham S, Trott DJ. 2020. Genomic analysis of phylogenetic group B2 extraintestinal pathogenic E. coli causing infections in dogs in Australia. Vet Microbiol 248:108783. doi: 10.1016/j.vetmic.2020.108783 - DOI - PubMed
MeSH terms
Substances
Grants and funding
- 201866/WT_/Wellcome Trust/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- 001/Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES)
- 2022/11917-1/Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP)
- 306396/2020-3/Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq)
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

