Recognition-Encoded Molecules: A Minimal Self-Replicator
- PMID: 39235971
- PMCID: PMC11632402
- DOI: 10.1002/chem.202401667
Recognition-Encoded Molecules: A Minimal Self-Replicator
Abstract
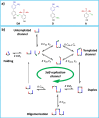
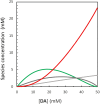
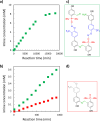
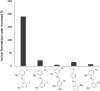
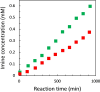
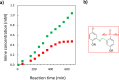
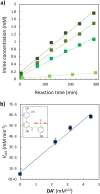
Nucleic acids, with their unique duplex structure, which is key for information replication, have sparked interest in self-replication's role in life's origins. Early template-based replicators, initially built on short oligonucleotides, expanded to include peptides and synthetic molecules. We explore here the potential of a class of synthetic duplex-forming oligoanilines, as self-replicators. We have recently developed oligoanilines equipped with 2-trifluoromethylphenol-phosphine oxide H-bond base pairs and we investigate whether the imine formed between aniline and aldehyde complementary monomers can self-replicate. Despite lacking a clear sigmoidal kinetic profile, control experiments with a methylated donor and a competitive inhibitor support self-replication. Further investigations with the reduced aniline dimer demonstrate templated synthesis, revealing a characteristic parabolic growth. After showing sequence selective duplex formation, templated synthesis and the emergence of catalytic function, the self-replication behaviour further suggests that the unique properties of nucleic acids can be paralleled by synthetic recognition-encoded molecules.
Keywords: Duplex; H-bonding; Oligoaniline; Recognition; Self-replicator.
© 2024 The Author(s). Chemistry - A European Journal published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Kiedrowski G. V., Angew. Chem. 1986, 98, 932–934.
-
- Appel R., Niemann B., Schuhn W., Knoch F., Angew. Chem. Int. Ed. Eng. 1986, 25, 932–932.
-
- Zielinski W. S., Orgel L. E., Nature 1987, 327, 346–347. - PubMed
-
- Lee D. H., Granja J. R., Martinez J. A., Severin K., Ghadiri M. R., Nature 1996, 382, 525–528. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

