MVST: Identifying spatial domains of spatial transcriptomes from multiple views using multi-view graph convolutional networks
- PMID: 39235988
- PMCID: PMC11376559
- DOI: 10.1371/journal.pcbi.1012409
MVST: Identifying spatial domains of spatial transcriptomes from multiple views using multi-view graph convolutional networks
Abstract
Spatial transcriptome technology can parse transcriptomic data at the spatial level to detect high-throughput gene expression and preserve information regarding the spatial structure of tissues. Identifying spatial domains, that is identifying regions with similarities in gene expression and histology, is the most basic and critical aspect of spatial transcriptome data analysis. Most current methods identify spatial domains only through a single view, which may obscure certain important information and thus fail to make full use of the information embedded in spatial transcriptome data. Therefore, we propose an unsupervised clustering framework based on multiview graph convolutional networks (MVST) to achieve accurate spatial domain recognition by the learning graph embedding features of neighborhood graphs constructed from gene expression information, spatial location information, and histopathological image information through multiview graph convolutional networks. By exploring spatial transcriptomes from multiple views, MVST enables data from all parts of the spatial transcriptome to be comprehensively and fully utilized to obtain more accurate spatial expression patterns. We verified the effectiveness of MVST on real spatial transcriptome datasets, the robustness of MVST on some simulated datasets, and the reasonableness of the framework structure of MVST in ablation experiments, and from the experimental results, it is clear that MVST can achieve a more accurate spatial domain identification compared with the current more advanced methods. In conclusion, MVST is a powerful tool for spatial transcriptome research with improved spatial domain recognition.
Copyright: © 2024 Duan et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


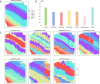
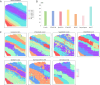

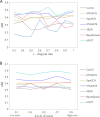
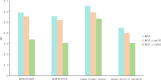
References
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

