Unveiling inter-embryo variability in spindle length over time: Towards quantitative phenotype analysis
- PMID: 39236069
- PMCID: PMC11376571
- DOI: 10.1371/journal.pcbi.1012330
Unveiling inter-embryo variability in spindle length over time: Towards quantitative phenotype analysis
Abstract
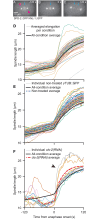
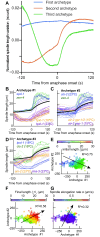
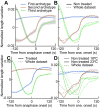
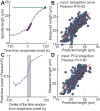
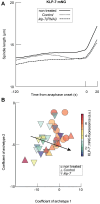
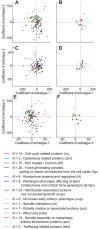
How can inter-individual variability be quantified? Measuring many features per experiment raises the question of choosing them to recapitulate high-dimensional data. Tackling this challenge on spindle elongation phenotypes, we showed that only three typical elongation patterns describe spindle elongation in C. elegans one-cell embryo. These archetypes, automatically extracted from the experimental data using principal component analysis (PCA), accounted for more than 95% of inter-individual variability of more than 1600 experiments across more than 100 different conditions. The two first archetypes were related to spindle average length and anaphasic elongation rate. The third archetype, accounting for 6% of the variability, was novel and corresponded to a transient spindle shortening in late metaphase, reminiscent of kinetochore function-defect phenotypes. Importantly, these three archetypes were robust to the choice of the dataset and were found even considering only non-treated conditions. Thus, the inter-individual differences between genetically perturbed embryos have the same underlying nature as natural inter-individual differences between wild-type embryos, independently of the temperatures. We thus propose that beyond the apparent complexity of the spindle, only three independent mechanisms account for spindle elongation, weighted differently in the various conditions. Interestingly, the spindle-length archetypes covered both metaphase and anaphase, suggesting that spindle elongation in late metaphase is sufficient to predict the late anaphase length. We validated this idea using a machine-learning approach. Finally, given amounts of these three archetypes could represent a quantitative phenotype. To take advantage of this, we set out to predict interacting genes from a seed based on the PCA coefficients. We exemplified this firstly on the role of tpxl-1 whose homolog tpx2 is involved in spindle microtubule branching, secondly the mechanism regulating metaphase length, and thirdly the central spindle players which set the length at anaphase. We found novel interactors not in public databases but supported by recent experimental publications.
Copyright: © 2024 Le Cunff et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

