Dual-role epitope on SARS-CoV-2 spike enhances and neutralizes viral entry across different variants
- PMID: 39236072
- PMCID: PMC11407660
- DOI: 10.1371/journal.ppat.1012493
Dual-role epitope on SARS-CoV-2 spike enhances and neutralizes viral entry across different variants
Abstract
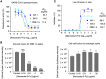
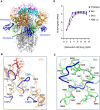
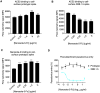
Grasping the roles of epitopes in viral glycoproteins is essential for unraveling the structure and function of these proteins. Up to now, all identified epitopes have been found to either neutralize, have no effect on, or enhance viral entry into cells. Here, we used nanobodies (single-domain antibodies) as probes to investigate a unique epitope on the SARS-CoV-2 spike protein, located outside the protein's receptor-binding domain. Nanobody binding to this epitope enhances the cell entry of prototypic SARS-CoV-2, while neutralizing the cell entry of SARS-CoV-2 Omicron variant. Moreover, nanobody binding to this epitope promotes both receptor binding activity and post-attachment activity of prototypic spike, explaining the enhanced viral entry. The opposite occurs with Omicron spike, explaining the neutralized viral entry. This study reveals a unique epitope that can both enhance and neutralize viral entry across distinct viral variants, suggesting that epitopes may vary their roles depending on the viral context. Consequently, antibody therapies should be assessed across different viral variants to confirm their efficacy and safety.
Copyright: © 2024 Ye et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The University of Minnesota has filed a patent on Nanosota-5, -6, and -7 with F.L., G.Y., F.B., and B.L. as inventors.
Figures


References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

