pyPAGE: A framework for Addressing biases in gene-set enrichment analysis-A case study on Alzheimer's disease
- PMID: 39236079
- PMCID: PMC11421795
- DOI: 10.1371/journal.pcbi.1012346
pyPAGE: A framework for Addressing biases in gene-set enrichment analysis-A case study on Alzheimer's disease
Abstract
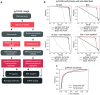
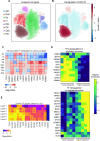
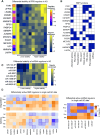
Inferring the driving regulatory programs from comparative analysis of gene expression data is a cornerstone of systems biology. Many computational frameworks were developed to address this problem, including our iPAGE (information-theoretic Pathway Analysis of Gene Expression) toolset that uses information theory to detect non-random patterns of expression associated with given pathways or regulons. Our recent observations, however, indicate that existing approaches are susceptible to the technical biases that are inherent to most real world annotations. To address this, we have extended our information-theoretic framework to account for specific biases and artifacts in biological networks using the concept of conditional information. To showcase pyPAGE, we performed a comprehensive analysis of regulatory perturbations that underlie the molecular etiology of Alzheimer's disease (AD). pyPAGE successfully recapitulated several known AD-associated gene expression programs. We also discovered several additional regulons whose differential activity is significantly associated with AD. We further explored how these regulators relate to pathological processes in AD through cell-type specific analysis of single cell and spatial gene expression datasets. Our findings showcase the utility of pyPAGE as a precise and reliable biomarker discovery in complex diseases such as Alzheimer's disease.
Copyright: © 2024 Bakulin et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures