Environmental modulators of algae-bacteria interactions at scale
- PMID: 39236710
- PMCID: PMC11412779
- DOI: 10.1016/j.cels.2024.08.002
Environmental modulators of algae-bacteria interactions at scale
Abstract
Interactions between photosynthetic and heterotrophic microbes play a key role in global primary production. Understanding phototroph-heterotroph interactions remains challenging because these microbes reside in chemically complex environments. Here, we leverage a massively parallel droplet microfluidic platform that enables us to interrogate interactions between photosynthetic algae and heterotrophic bacteria in >100,000 communities across ∼525 environmental conditions with varying pH, carbon availability, and phosphorus availability. By developing a statistical framework to dissect interactions in this complex dataset, we reveal that the dependence of algae-bacteria interactions on nutrient availability is strongly modulated by pH and buffering capacity. Furthermore, we show that the chemical identity of the available organic carbon source controls how pH, buffering capacity, and nutrient availability modulate algae-bacteria interactions. Our study reveals the previously underappreciated role of pH in modulating phototroph-heterotroph interactions and provides a framework for thinking about interactions between phototrophs and heterotrophs in more natural contexts.
Keywords: algae-bacteria interactions; buffering capacity; ecological interactions; high-throughput microfluidic platform; pH; phototroph-heterotroph interactions.
Copyright © 2024 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests C.G. is currently employed by the Department of Civil and Environmental Engineering, Massachusetts Institute of Technology. Z.L. is currently employed by BillionToOne Inc.
Figures

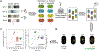

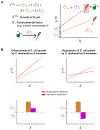


References
-
- Mickalide H, and Kuehn S (2019). Higher-Order Interaction between Species Inhibits Bacterial Invasion of a Phototroph-Predator Microbial Community. Cell Systems 9, 521–533.e10. - PubMed
-
- Thompson AW, Foster RA, Krupke A, Carter BJ, Musat N, Vaulot D, Kuypers MMM, and Zehr JP (2012). Unicellular cyanobacterium symbiotic with a single-celled eukaryotic alga. Science 337, 1546–1550. - PubMed
-
- Blanton LV, Charbonneau MR, Salih T, Barratt MJ, Venkatesh S, Ilkaveya O, Subramanian S, Manary MJ, Trehan I, Jorgensen JM, Fan Y-M, Henrissat B, Leyn SA, Rodionov DA, Osterman AL, Maleta KM, Newgard CB, Ashorn P, Dewey KG, and Gordon JI (2016). Gut bacteria that prevent growth impairments transmitted by microbiota from malnourished children. Science 351, aad3311. - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

