Discovery of therapeutic targets in cancer using chromatin accessibility and transcriptomic data
- PMID: 39236711
- PMCID: PMC11415227
- DOI: 10.1016/j.cels.2024.08.004
Discovery of therapeutic targets in cancer using chromatin accessibility and transcriptomic data
Abstract
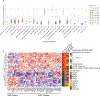
Most cancer types lack targeted therapeutic options, and when first-line targeted therapies are available, treatment resistance is a huge challenge. Recent technological advances enable the use of assay for transposase-accessible chromatin with sequencing (ATAC-seq) and RNA sequencing (RNA-seq) on patient tissue in a high-throughput manner. Here, we present a computational approach that leverages these datasets to identify drug targets based on tumor lineage. We constructed gene regulatory networks for 371 patients of 22 cancer types using machine learning approaches trained with three-dimensional genomic data for enhancer-to-promoter contacts. Next, we identified the key transcription factors (TFs) in these networks, which are used to find therapeutic vulnerabilities, by direct targeting of either TFs or the proteins that they interact with. We validated four candidates identified for neuroendocrine, liver, and renal cancers, which have a dismal prognosis with current therapeutic options.
Keywords: ATAC-seq; cancer; drug repurposing; functional genomics; networks; regulatory networks; therapy.
Copyright © 2024 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures
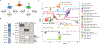



References
-
- Fisher B, Costantino JP, Wickerham DL, Redmond CK, Kavanah M, Cronin WM, Vogel V, Robidoux A, Dimitrov N, Atkins J, et al. (1998). Tamoxifen for Prevention of Breast Cancer: Report of the National Surgical Adjuvant Breast and Bowel Project P-1 Study. JNCI: Journal of the National Cancer Institute 90, 1371–1388. 10.1093/jnci/90.18.1371. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

