Combinatorial targeting of glutamine metabolism and lysosomal-based lipid metabolism effectively suppresses glioblastoma
- PMID: 39236712
- PMCID: PMC11524980
- DOI: 10.1016/j.xcrm.2024.101706
Combinatorial targeting of glutamine metabolism and lysosomal-based lipid metabolism effectively suppresses glioblastoma
Abstract
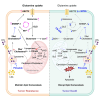
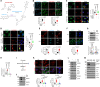
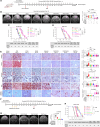
Antipsychotic drugs have been shown to have antitumor effects but have had limited potency in the clinic. Here, we unveil that pimozide inhibits lysosome hydrolytic function to suppress fatty acid and cholesterol release in glioblastoma (GBM), the most lethal brain tumor. Unexpectedly, GBM develops resistance to pimozide by boosting glutamine consumption and lipogenesis. These elevations are driven by SREBP-1, which we find upregulates the expression of ASCT2, a key glutamine transporter. Glutamine, in turn, intensifies SREBP-1 activation through the release of ammonia, creating a feedforward loop that amplifies both glutamine metabolism and lipid synthesis, leading to drug resistance. Disrupting this loop via pharmacological targeting of ASCT2 or glutaminase, in combination with pimozide, induces remarkable mitochondrial damage and oxidative stress, leading to GBM cell death in vitro and in vivo. Our findings underscore the promising therapeutic potential of effectively targeting GBM by combining glutamine metabolism inhibition with lysosome suppression.
Keywords: ASCT2; GLS; SREBP-1; cholesterol; fatty acids; glioblastoma; glutamine; lipid droplets; lysosome; pimozide.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures





References
-
- Bagley S.J., Kothari S., Rahman R., Lee E.Q., Dunn G.P., Galanis E., Chang S.M., Nabors L.B., Ahluwalia M.S., Stupp R., et al. Glioblastoma Clinical Trials: Current Landscape and Opportunities for Improvement. Clin. Cancer Res. 2022;28:594–602. doi: 10.1158/1078-0432.CCR-21-2750. - DOI - PMC - PubMed
-
- Matteoni S., Matarrese P., Ascione B., Buccarelli M., Ricci-Vitiani L., Pallini R., Villani V., Pace A., Paggi M.G., Abbruzzese C. Anticancer Properties of the Antipsychotic Drug Chlorpromazine and Its Synergism With Temozolomide in Restraining Human Glioblastoma Proliferation In Vitro. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.635472. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

