A robust mouse model of HPIV-3 infection and efficacy of GS-441524 against virus-induced lung pathology
- PMID: 39237507
- PMCID: PMC11377736
- DOI: 10.1038/s41467-024-52071-5
A robust mouse model of HPIV-3 infection and efficacy of GS-441524 against virus-induced lung pathology
Abstract
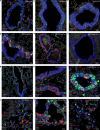
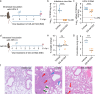
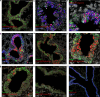
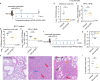
Human parainfluenza virus type 3 (HPIV-3) can cause severe respiratory tract infections. There are no convenient small-animal infection models. Here, we show viral replication in the upper and lower airways of AG129 mice (double IFNα/β and IFNγ receptor knockout mice) upon intranasal inoculation. By multiplex fluorescence RNAscope and immunohistochemistry followed by confocal microscopy, we demonstrate viral tropism to ciliated cells and club cells of the bronchiolar epithelium. HPIV-3 causes a marked lung pathology. No virus transmission of the virus was observed by cohousing HPIV-3-infected AG129 mice with other mice. Oral treatment with GS-441524, the parent nucleoside of remdesivir, reduced infectious virus titers in the lung, with a relatively normal histology. Intranasal treatment also affords an antiviral effect. Thus, AG129 mice serve as a robust preclinical model for developing therapeutic and prophylactic strategies against HPIV-3. We suggest further investigation of GS-441524 and its prodrug forms to treat HPIV-3 infection in humans.
© 2024. The Author(s).
Conflict of interest statement
D.B. and J.B. are employees of Gilead Sciences which owns GS-441524. None of the other authors have any competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

