A new paradigm for generating high-quality cardiac pacemaker cells from mouse pluripotent stem cells
- PMID: 39237509
- PMCID: PMC11377569
- DOI: 10.1038/s41392-024-01942-w
A new paradigm for generating high-quality cardiac pacemaker cells from mouse pluripotent stem cells
Abstract
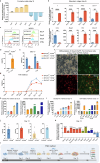
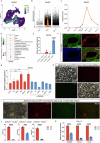
Cardiac biological pacing (BP) is one of the future directions for bradyarrhythmias intervention. Currently, cardiac pacemaker cells (PCs) used for cardiac BP are mainly derived from pluripotent stem cells (PSCs). However, the production of high-quality cardiac PCs from PSCs remains a challenge. Here, we developed a cardiac PC differentiation strategy by adopting dual PC markers and simulating the developmental route of PCs. First, two PC markers, Shox2 and Hcn4, were selected to establish Shox2:EGFP; Hcn4:mCherry mouse PSC reporter line. Then, by stepwise guiding naïve PSCs to cardiac PCs following naïve to formative pluripotency transition and manipulating signaling pathways during cardiac PCs differentiation, we designed the FSK method that increased the yield of SHOX2+; HCN4+ cells with typical PC characteristics, which was 12 and 42 folds higher than that of the embryoid body (EB) and the monolayer M10 methods respectively. In addition, the in vitro cardiac PCs differentiation trajectory was mapped by single-cell RNA sequencing (scRNA-seq), which resembled in vivo PCs development, and ZFP503 was verified as a key regulator of cardiac PCs differentiation. These PSC-derived cardiac PCs have the potential to drive advances in cardiac BP technology, help with the understanding of PCs (patho)physiology, and benefit drug discovery for PC-related diseases as well.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
- 82088101/National Natural Science Foundation of China (National Science Foundation of China)
- 81930013/National Natural Science Foundation of China (National Science Foundation of China)
- 82000377/National Natural Science Foundation of China (National Science Foundation of China)
- 81900297/National Natural Science Foundation of China (National Science Foundation of China)
- 82070338/National Natural Science Foundation of China (National Science Foundation of China)
LinkOut - more resources
Full Text Sources

