Immune landscape of oncohistone-mutant gliomas reveals diverse myeloid populations and tumor-promoting function
- PMID: 39237515
- PMCID: PMC11377583
- DOI: 10.1038/s41467-024-52096-w
Immune landscape of oncohistone-mutant gliomas reveals diverse myeloid populations and tumor-promoting function
Abstract
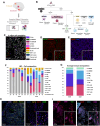
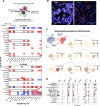
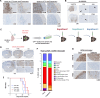
Histone H3-mutant gliomas are deadly brain tumors characterized by a dysregulated epigenome and stalled differentiation. In contrast to the extensive datasets available on tumor cells, limited information exists on their tumor microenvironment (TME), particularly the immune infiltrate. Here, we characterize the immune TME of H3.3K27M and G34R/V-mutant gliomas, and multiple H3.3K27M mouse models, using transcriptomic, proteomic and spatial single-cell approaches. Resolution of immune lineages indicates high infiltration of H3-mutant gliomas with diverse myeloid populations, high-level expression of immune checkpoint markers, and scarce lymphoid cells, findings uniformly reproduced in all H3.3K27M mouse models tested. We show these myeloid populations communicate with H3-mutant cells, mediating immunosuppression and sustaining tumor formation and maintenance. Dual inhibition of myeloid cells and immune checkpoint pathways show significant therapeutic benefits in pre-clinical syngeneic mouse models. Our findings provide a valuable characterization of the TME of oncohistone-mutant gliomas, and insight into the means for modulating the myeloid infiltrate for the benefit of patients.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

