RAD52 resolves transcription-replication conflicts to mitigate R-loop induced genome instability
- PMID: 39237529
- PMCID: PMC11377823
- DOI: 10.1038/s41467-024-51784-x
RAD52 resolves transcription-replication conflicts to mitigate R-loop induced genome instability
Abstract
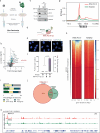
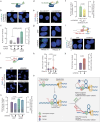
Collisions of the transcription and replication machineries on the same DNA strand can pose a significant threat to genomic stability. These collisions occur in part due to the formation of RNA-DNA hybrids termed R-loops, in which a newly transcribed RNA molecule hybridizes with the DNA template strand. This study investigated the role of RAD52, a known DNA repair factor, in preventing collisions by directing R-loop formation and resolution. We show that RAD52 deficiency increases R-loop accumulation, exacerbating collisions and resulting in elevated DNA damage. Furthermore, RAD52's ability to interact with the transcription machinery, coupled with its capacity to facilitate R-loop dissolution, highlights its role in preventing collisions. Lastly, we provide evidence of an increased mutational burden from double-strand breaks at conserved R-loop sites in human tumor samples, which is increased in tumors with low RAD52 expression. In summary, this study underscores the importance of RAD52 in orchestrating the balance between replication and transcription processes to prevent collisions and maintain genome stability.
© 2024. The Author(s).
Conflict of interest statement
S.F.B. owns equity in, receives compensation from, and serves as a consultant for and serves on the Scientific Advisory Board (SAB) and Board of Directors (BOD) of Volastra Therapeutics Inc and he serves on the scientific advisory board of Meliora Therapeutics. J.S.R.-F. reports current employment at AstraZeneca and stocks in AstraZeneca, Repare Therapeutics, Paige.AI; J.S.R.-F. previously held a fiduciary role in Grupo Oncoclinicas and was a consultant with Goldman Sachs Merchant Banking, Bain Capital, Repare Therapeutics, Paige.AI, Volition Rx and MultiplexDx. S.N.P. reports consulting fees from the following companies outside the scope of this study: Varian Medical Systems, Philips Healthcare and AstraZeneca, as well as research funding from Philips Healthcare and Varian Medical Systems. The remaining authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

