Quantifying DNA damage following light sheet and confocal imaging of the mammalian embryo
- PMID: 39237572
- PMCID: PMC11377761
- DOI: 10.1038/s41598-024-71443-x
Quantifying DNA damage following light sheet and confocal imaging of the mammalian embryo
Abstract
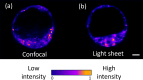
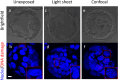
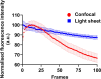
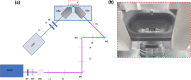
Embryo quality assessment by optical imaging is increasing in popularity. Among available optical techniques, light sheet microscopy has emerged as a superior alternative to confocal microscopy due to its geometry, enabling faster image acquisition with reduced photodamage to the sample. However, previous assessments of photodamage induced by imaging may have failed to measure more subtle impacts. In this study, we employed DNA damage as a sensitive indicator of photodamage. We use light sheet microscopy with excitation at a wavelength of 405 nm for imaging embryo autofluorescence and compare its performance to laser scanning confocal microscopy. At an equivalent signal-to-noise ratio for images acquired with both modalities, light sheet microscopy reduced image acquisition time by ten-fold, and did not induce DNA damage when compared to non-imaged embryos. In contrast, imaging with confocal microscopy led to significantly higher levels of DNA damage within embryos and had a higher photobleaching rate. Light sheet imaging is also capable of inducing DNA damage within the embryo but requires multiple cycles of volumetric imaging. Collectively, this study confirms that light sheet microscopy is faster and safer than confocal microscopy for imaging live embryos, indicating its potential as a label-free diagnostic for embryo quality.
© 2024. The Author(s).
Figures



References
-
- Adolfsson, E. & Andershed, A. N. Morphology vs morphokinetics: A retrospective comparison of inter-observer and intra-observer agreement between embryologists on blastocysts with known implantation outcome. JBRA Assist. Reprod.22, 228–237. 10.5935/1518-0557.20180042 (2018). 10.5935/1518-0557.20180042 - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

