Prion protein alters viral control and enhances pathology after perinatal cytomegalovirus infection
- PMID: 39237588
- PMCID: PMC11377837
- DOI: 10.1038/s41467-024-51931-4
Prion protein alters viral control and enhances pathology after perinatal cytomegalovirus infection
Abstract
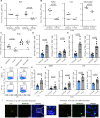
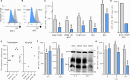
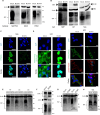
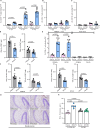
Cytomegalovirus (CMV) infection poses risks to newborns, necessitating effective therapies. Given that the damage includes both viral infection of brain cells and immune system-related damage, here we investigate the involvement of cellular prion protein (PrP), which plays vital roles in neuroprotection and immune regulation. Using a murine model, we show the role of PrP in tempering neonatal T cell immunity during CMV infection. PrP-null mice exhibit enhanced viral control through elevated virus-specific CD8 T cell responses, leading to reduced viral titers and pathology. We further unravel the molecular mechanisms by showing CMV-induced upregulation followed by release of PrP via the metalloproteinase ADAM10, impairing CD8 T cell response specifically in neonates. Additionally, we confirm PrP downregulation in human CMV (HCMV)-infected fibroblasts, underscoring the broader relevance of our observations beyond the murine model. Furthermore, our study highlights how PrP, under the stress of viral pathogenesis, reveals its impact on neonatal immune modulation.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

