A Gaussia luciferase reporter assay for the evaluation of coronavirus Nsp5/3CLpro activity
- PMID: 39237598
- PMCID: PMC11377810
- DOI: 10.1038/s41598-024-71305-6
A Gaussia luciferase reporter assay for the evaluation of coronavirus Nsp5/3CLpro activity
Abstract
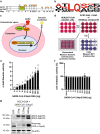
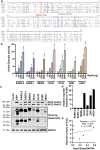
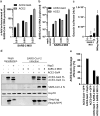
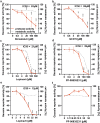
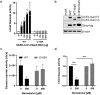
Human coronaviruses (hCoVs) infect millions of people every year. Among these, MERS, SARS-CoV-1, and SARS-CoV-2 caused significant morbidity and mortality and their emergence highlights the risk of possible future coronavirus outbreaks. Therefore, broadly-active anti-coronavirus drugs are needed. Pharmacological inhibition of the hCoV protease Nsp5 (3CLpro) is clinically beneficial as shown by the wide and effective use of Paxlovid (nirmatrelvir, ritonavir). However, further treatment options are required due to the risk of drug resistance. To facilitate the assessment of coronavirus protease function and its pharmacological inhibition, we developed an assay allowing rapid and reliable quantification of Nsp5 activity under biosafety level 1 conditions. It is based on an ACE2-Gal4 transcription factor fusion protein separated by a Nsp5 recognition site. Cleavage by Nsp5 releases the Gal4 transcription factor, which then induces the expression of Gaussia luciferase. Our assay is compatible with Nsp5 proteases from all hCoVs and allows simultaneous measurement of inhibitory and cytotoxic effects of the tested compounds. Proof-of-concept measurements confirmed that nirmatrelvir, GC376 and lopinavir inhibit SARS-CoV-2 Nsp5 function. Furthermore, the assay accurately predicted the impact of Nsp5 mutations on catalytic activity and inhibitor sensitivity. Overall, the reporter assay is suitable for evaluating viral protease activity.
Keywords: 3CLpro; Coronaviruses; Gaussia reporter assay; Nsp5; Protease inhibitor; SARS-CoV-2.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Detecting SARS-CoV-2 3CLpro expression and activity using a polyclonal antiserum and a luciferase-based biosensor.Virology. 2021 Apr;556:73-78. doi: 10.1016/j.virol.2021.01.010. Epub 2021 Jan 26. Virology. 2021. PMID: 33548599 Free PMC article.
-
Development of a Cell-Based Luciferase Complementation Assay for Identification of SARS-CoV-2 3CLpro Inhibitors.Viruses. 2021 Jan 24;13(2):173. doi: 10.3390/v13020173. Viruses. 2021. PMID: 33498923 Free PMC article.
-
A highly sensitive cell-based luciferase assay for high-throughput automated screening of SARS-CoV-2 nsp5/3CLpro inhibitors.Antiviral Res. 2022 May;201:105272. doi: 10.1016/j.antiviral.2022.105272. Epub 2022 Mar 9. Antiviral Res. 2022. PMID: 35278581 Free PMC article.
-
Targeting novel structural and functional features of coronavirus protease nsp5 (3CLpro, Mpro) in the age of COVID-19.J Gen Virol. 2021 Mar;102(3):001558. doi: 10.1099/jgv.0.001558. Epub 2021 Jan 28. J Gen Virol. 2021. PMID: 33507143 Free PMC article. Review.
-
Recent Advances in SARS-CoV-2 Main Protease Inhibitors: From Nirmatrelvir to Future Perspectives.Biomolecules. 2023 Sep 2;13(9):1339. doi: 10.3390/biom13091339. Biomolecules. 2023. PMID: 37759739 Free PMC article. Review.
References
-
- Coronavirus disease (COVID-19). https://www.who.int/news-room/fact-sheets/detail/coronavirus-disease-(co... (Accessed 18 April 2024).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

