Temporal stability of the rumen microbiome and its longitudinal associations with performance traits in beef cattle
- PMID: 39237607
- PMCID: PMC11377694
- DOI: 10.1038/s41598-024-70770-3
Temporal stability of the rumen microbiome and its longitudinal associations with performance traits in beef cattle
Abstract
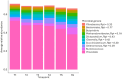
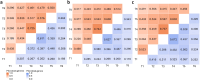
The rumen microbiome is the focus of a growing body of research, mostly based on investigation of rumen fluid samples collected once from each animal. Exploring the temporal stability of rumen microbiome profiles is imperative, as it enables evaluating the reliability of findings obtained through single-timepoint sampling. We explored the temporal stability of rumen microbiomes considering taxonomic and functional aspects across the 7-month growing-finishing phase spanning 6 timepoints. We identified a temporally stable core microbiome, encompassing 515 microbial genera (e.g., Methanobacterium) and 417 microbial KEGG genes (e.g., K00856-adenosine kinase). The temporally stable core microbiome profiles collected from all timepoints were strongly associated with production traits with substantial economic and environmental impact (e.g., average daily gain, daily feed intake, and methane emissions); 515 microbial genera explained 45-83%, and 417 microbial genes explained 44-83% of their phenotypic variation. Microbiome profiles influenced by the bovine genome explained 54-87% of the genetic variation of bovine traits. Overall, our results provide evidence that the temporally stable core microbiome identified can accurately predict host performance traits at phenotypic and genetic level based on a single timepoint sample taken as early as 7 months prior to slaughter.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Gruninger, R. J., Ribeiro, G. O., Cameron, A. & McAllister, T. A. Invited review: Application of meta-omics to understand the dynamic nature of the rumen microbiome and how it responds to diet in ruminants. Animal13, 1843–1854. 10.1017/S1751731119000752 (2019). 10.1017/S1751731119000752 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

