A bitopic agonist bound to the dopamine 3 receptor reveals a selectivity site
- PMID: 39237617
- PMCID: PMC11377762
- DOI: 10.1038/s41467-024-51993-4
A bitopic agonist bound to the dopamine 3 receptor reveals a selectivity site
Abstract
Although aminergic GPCRs are the target for ~25% of approved drugs, developing subtype selective drugs is a major challenge due to the high sequence conservation at their orthosteric binding site. Bitopic ligands are covalently joined orthosteric and allosteric pharmacophores with the potential to boost receptor selectivity and improve current medications by reducing off-target side effects. However, the lack of structural information on their binding mode impedes rational design. Here we determine the cryo-EM structure of the hD3R:GαOβγ complex bound to the D3R selective bitopic agonist FOB02-04A. Structural, functional and computational analyses provide insights into its binding mode and point to a new TM2-ECL1-TM1 region, which requires the N-terminal ordering of TM1, as a major determinant of subtype selectivity in aminergic GPCRs. This region is underexploited in drug development, expands the established secondary binding pocket in aminergic GPCRs and could potentially be used to design novel and subtype selective drugs.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
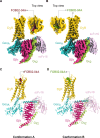
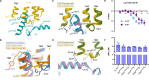



Update of
-
Structure of the dopamine D3 receptor bound to a bitopic agonist reveals a new specificity site in an expanded allosteric pocket.Res Sq [Preprint]. 2023 Dec 19:rs.3.rs-3433207. doi: 10.21203/rs.3.rs-3433207/v1. Res Sq. 2023. Update in: Nat Commun. 2024 Sep 5;15(1):7759. doi: 10.1038/s41467-024-51993-4. PMID: 38196573 Free PMC article. Updated. Preprint.
Similar articles
-
Structure of the dopamine D3 receptor bound to a bitopic agonist reveals a new specificity site in an expanded allosteric pocket.Res Sq [Preprint]. 2023 Dec 19:rs.3.rs-3433207. doi: 10.21203/rs.3.rs-3433207/v1. Res Sq. 2023. Update in: Nat Commun. 2024 Sep 5;15(1):7759. doi: 10.1038/s41467-024-51993-4. PMID: 38196573 Free PMC article. Updated. Preprint.
-
Prescription of Controlled Substances: Benefits and Risks.2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. 2025 Jul 6. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025 Jan–. PMID: 30726003 Free Books & Documents.
-
Structure-based discovery of positive allosteric modulators of the A1 adenosine receptor.Proc Natl Acad Sci U S A. 2025 Jul 15;122(28):e2421687122. doi: 10.1073/pnas.2421687122. Epub 2025 Jul 7. Proc Natl Acad Sci U S A. 2025. PMID: 40623180 Free PMC article.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Management of urinary stones by experts in stone disease (ESD 2025).Arch Ital Urol Androl. 2025 Jun 30;97(2):14085. doi: 10.4081/aiua.2025.14085. Epub 2025 Jun 30. Arch Ital Urol Androl. 2025. PMID: 40583613 Review.
Cited by
-
2-Phenylcyclopropylmethylamine (PCPMA) Derivatives as D3R-Selective Ligands for 3D-QSAR, Docking and Molecular Dynamics Simulation Studies.Int J Mol Sci. 2025 Apr 10;26(8):3559. doi: 10.3390/ijms26083559. Int J Mol Sci. 2025. PMID: 40332026 Free PMC article.
-
Go/z-biased coupling profile of the dopamine D3 receptor.bioRxiv [Preprint]. 2025 Aug 12:2025.08.08.668522. doi: 10.1101/2025.08.08.668522. bioRxiv. 2025. PMID: 40832361 Free PMC article. Preprint.
-
AI meets physics in computational structure-based drug discovery for GPCRs.NPJ Drug Discov. 2025;2(1):16. doi: 10.1038/s44386-025-00019-0. Epub 2025 Jul 3. NPJ Drug Discov. 2025. PMID: 40621551 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

