Conditional protein splicing of the Mycobacterium tuberculosis RecA intein in its native host
- PMID: 39237639
- PMCID: PMC11377839
- DOI: 10.1038/s41598-024-71248-y
Conditional protein splicing of the Mycobacterium tuberculosis RecA intein in its native host
Abstract
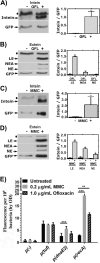
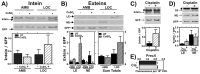
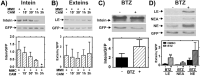
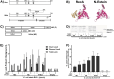
The recA gene, encoding Recombinase A (RecA) is one of three Mycobacterium tuberculosis (Mtb) genes encoding an in-frame intervening protein sequence (intein) that must splice out of precursor host protein to produce functional protein. Ongoing debate about whether inteins function solely as selfish genetic elements or benefit their host cells requires understanding of interplay between inteins and their hosts. We measured environmental effects on native RecA intein splicing within Mtb using a combination of western blots and promoter reporter assays. RecA splicing was stimulated in bacteria exposed to DNA damaging agents or by treatment with copper in hypoxic, but not normoxic, conditions. Spliced RecA was processed by the Mtb proteasome, while free intein was degraded efficiently by other unknown mechanisms. Unspliced precursor protein was not observed within Mtb despite its accumulation during ectopic expression of Mtb recA within E. coli. Surprisingly, Mtb produced free N-extein in some conditions, and ectopic expression of Mtb N-extein activated LexA in E. coli. These results demonstrate that the bacterial environment greatly impacts RecA splicing in Mtb, underscoring the importance of studying intein splicing in native host environments and raising the exciting possibility of intein splicing as a novel regulatory mechanism in Mtb.
Keywords: DNA damage repair; Exaptation; Gene expression; Mobile elements; Post-translational gene regulation; SOS response.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Update of
-
Conditional protein splicing of the Mycobacterium tuberculosis RecA intein in its native host.bioRxiv [Preprint]. 2024 Apr 15:2024.04.15.589443. doi: 10.1101/2024.04.15.589443. bioRxiv. 2024. Update in: Sci Rep. 2024 Sep 5;14(1):20664. doi: 10.1038/s41598-024-71248-y. PMID: 38659745 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

