Biological function and potential application of PANoptosis-related genes in colorectal carcinogenesis
- PMID: 39237645
- PMCID: PMC11377449
- DOI: 10.1038/s41598-024-71625-7
Biological function and potential application of PANoptosis-related genes in colorectal carcinogenesis
Abstract
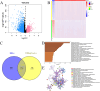
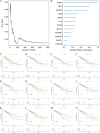
PANoptosis induces programmed cell death (PCD) through extensive crosstalk and is associated with development of cancer. However, the functional mechanisms, clinical significance, and potential applications of PANoptosis-related genes (PRGs) in colorectal cancer (CRC) have not been fully elucidated. Functional enrichment of key PRGs was analyzed based on databases, and relationships between key PRGs and the immune microenvironment, immune cell infiltration, chemotherapy drug sensitivity, tumor progression genes, single-cell cellular subgroups, signal transduction pathways, transcription factor regulation, and miRNA regulatory networks were systematically explored. This study identified 5 key PRGs associated with CRC: BCL10, CDKN2A, DAPK1, PYGM and TIMP1. Then, RT-PCR was used to verify expression of these genes in CRC cells and tissues. Clinical significance and prognostic value of key genes were further verified by multiple datasets. Analyses of the immune microenvironment, immune cell infiltration, chemotherapy drug sensitivity, tumor progression genes, single-cell cellular subgroups, and signal transduction pathways suggest a close relationship between these key genes and development of CRC. In addition, a novel prognostic nomogram model for CRC was successfully constructed by combining important clinical indicators and the key genes. In conclusion, our findings offer new insights for understanding the pathogenesis of CRC, predicting CRC prognosis, and identifying multiple therapeutic targets for future CRC therapy.
Keywords: Chemotherapy drug sensitivity; Colorectal cancer; Immune microenvironment; Nomogram; PANoptosis; Prognosis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

