PPM1D activity promotes cellular transformation by preventing senescence and cell death
- PMID: 39237765
- PMCID: PMC11473410
- DOI: 10.1038/s41388-024-03149-3
PPM1D activity promotes cellular transformation by preventing senescence and cell death
Abstract
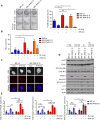
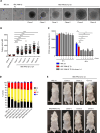
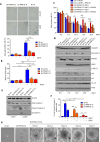
Cell cycle checkpoints, oncogene-induced senescence and programmed cell death represent intrinsic barriers to tumorigenesis. Protein phosphatase magnesium-dependent 1 (PPM1D) is a negative regulator of the tumour suppressor p53 and has been implicated in termination of the DNA damage response. Here, we addressed the consequences of increased PPM1D activity resulting from the gain-of-function truncating mutations in exon 6 of the PPM1D. We show that while control cells permanently exit the cell cycle and reside in senescence in the presence of DNA damage caused by ionising radiation or replication stress induced by the active RAS oncogene, RPE1-hTERT and BJ-hTERT cells carrying the truncated PPM1D continue proliferation in the presence of DNA damage, form micronuclei and accumulate genomic rearrangements revealed by karyotyping. Further, we show that increased PPM1D activity promotes cell growth in the soft agar and formation of tumours in xenograft models. Finally, expression profiling of the transformed clones revealed dysregulation of several oncogenic and tumour suppressor pathways. Our data support the oncogenic potential of PPM1D in the context of exposure to ionising radiation and oncogene-induced replication stress.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. - PubMed
-
- Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–70. - PubMed
-
- Collado M, Gil J, Efeyan A, Guerra C, Schuhmacher AJ, Barradas M. et al.Tumour biology: senescence in premalignant tumours.Nature.2005;436:642 10.1038/436642a. - PubMed
-
- Braig M, Lee S, Loddenkemper C, Rudolph C, Peters AH, Schlegelberger B, et al. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005;436:660–5. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

