Mapping protein-DNA interactions with DiMeLo-seq
- PMID: 39237830
- PMCID: PMC11674881
- DOI: 10.1038/s41596-024-01032-9
Mapping protein-DNA interactions with DiMeLo-seq
Abstract
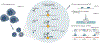
We recently developed directed methylation with long-read sequencing (DiMeLo-seq) to map protein-DNA interactions genome wide. DiMeLo-seq is capable of mapping multiple interaction sites on single DNA molecules, profiling protein binding in the context of endogenous DNA methylation, identifying haplotype-specific protein-DNA interactions and mapping protein-DNA interactions in repetitive regions of the genome that are difficult to study with short-read methods. With DiMeLo-seq, adenines in the vicinity of a protein of interest are methylated in situ by tethering the Hia5 methyltransferase to an antibody using protein A. Protein-DNA interactions are then detected by direct readout of adenine methylation with long-read, single-molecule DNA sequencing platforms such as Nanopore sequencing. Here we present a detailed protocol and practical guidance for performing DiMeLo-seq. This protocol can be run on nuclei from fresh, lightly fixed or frozen cells. The protocol requires 1-2 d for performing in situ targeted methylation, 1-5 d for library preparation depending on desired fragment length and 1-3 d for Nanopore sequencing depending on desired sequencing depth. The protocol requires basic molecular biology skills and equipment, as well as access to a Nanopore sequencer. We also provide a Python package, dimelo, for analysis of DiMeLo-seq data.
© 2024. Springer Nature Limited.
Conflict of interest statement
Competing interests: N.A., A.M., K.S., A.F.S. and A.S. are co-inventors on a patent application related to this work. The remaining authors declare no competing interests.
Figures





References
-
- Robertson G. et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods 4, 651–657 (2007). - PubMed
-
- Johnson DS, Mortazavi A, Myers RM & Wold B Genome-wide mapping of in vivo protein–DNA interactions. Science 316, 1497–1502 (2007). - PubMed
-
- Barski A. et al. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 (2007). - PubMed
Publication types
MeSH terms
Substances
Associated data
- BioProject/PRJNA855257
- BioProject/PRJNA752170
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

