Antimicrobial activity of compounds identified by artificial intelligence discovery engine targeting enzymes involved in Neisseria gonorrhoeae peptidoglycan metabolism
- PMID: 39238057
- PMCID: PMC11375863
- DOI: 10.1186/s40659-024-00543-9
Antimicrobial activity of compounds identified by artificial intelligence discovery engine targeting enzymes involved in Neisseria gonorrhoeae peptidoglycan metabolism
Abstract
Background: Neisseria gonorrhoeae (Ng) causes the sexually transmitted disease gonorrhoea. There are no vaccines and infections are treated principally with antibiotics. However, gonococci rapidly develop resistance to every antibiotic class used and there is a need for developing new antimicrobial treatments. In this study we focused on two gonococcal enzymes as potential antimicrobial targets, namely the serine protease L,D-carboxypeptidase LdcA (NgO1274/NEIS1546) and the lytic transglycosylase LtgD (NgO0626/NEIS1212). To identify compounds that could interact with these enzymes as potential antimicrobials, we used the AtomNet virtual high-throughput screening technology. We then did a computational modelling study to examine the interactions of the most bioactive compounds with their target enzymes. The identified compounds were tested against gonococci to determine minimum inhibitory and bactericidal concentrations (MIC/MBC), specificity, and compound toxicity in vitro.
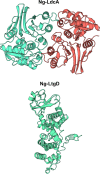
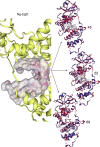
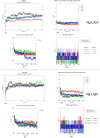
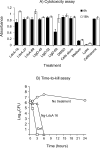
Results: AtomNet identified 74 compounds that could potentially interact with Ng-LdcA and 84 compounds that could potentially interact with Ng-LtgD. Through MIC and MBC assays, we selected the three best performing compounds for both enzymes. Compound 16 was the most active against Ng-LdcA, with a MIC50 value < 1.56 µM and MBC50/90 values between 0.195 and 0.39 µM. In general, the Ng-LdcA compounds showed higher activity than the compounds directed against Ng-LtgD, of which compound 45 had MIC50 values of 1.56-3.125 µM and MBC50/90 values between 3.125 and 6.25 µM. The compounds were specific for gonococci and did not kill other bacteria. They were also non-toxic for human conjunctival epithelial cells as judged by a resazurin assay. To support our biological data, in-depth computational modelling study detailed the interactions of the compounds with their target enzymes. Protein models were generated in silico and validated, the active binding sites and amino acids involved elucidated, and the interactions of the compounds interacting with the enzymes visualised through molecular docking and Molecular Dynamics Simulations for 50 ns and Molecular Mechanics Poisson-Boltzmann Surface Area (MM-PBSA).
Conclusions: We have identified bioactive compounds that appear to target the N. gonorrhoeae LdcA and LtgD enzymes. By using a reductionist approach involving biological and computational data, we propose that compound Ng-LdcA-16 and Ng-LtgD-45 are promising anti-gonococcal compounds for further development.
Keywords: Neisseria gonorrhoeae; Artificial intelligence; Bactericidal; Computational modelling; Peptidoglycan.
© 2024. The Author(s).
Conflict of interest statement
All authors declare that they have no competing interests.
Figures







References
-
- Mayor MT, Roett MA, Uduhiri KA. Diagnosis and management of gonococcal infections. Am Fam Phys. 2012;86(10):931–8. - PubMed
-
- Wetzler LM, Feavers IM, Gray-Owen SD, Jerse AE, Rice PA, Deal CD. Summary and recommendations from the National Institute of allergy and infectious diseases (NIAID) workshop “Gonorrhea vaccines: the way forward.” Clin Vacc Immunol. 2016;23(8):656–63. 10.1128/CVI.00230-16. 10.1128/CVI.00230-16 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

