Analysis of miRNAs involved in mouse brain injury upon Coxsackievirus A6 infection
- PMID: 39239635
- PMCID: PMC11374775
- DOI: 10.3389/fcimb.2024.1405689
Analysis of miRNAs involved in mouse brain injury upon Coxsackievirus A6 infection
Abstract
Introduction: Coxsackievirus A6 (CV-A6) has emerged as the predominant epidemic strain responsible for hand, foot and mouth disease (HFMD). CV-A6 infection can result in severe clinical manifestations, including encephalitis, meningitis, and potentially life-threatening central nervous system disorders. Our previous research findings demonstrated that neonatal mice infected with CV-A6 exhibited limb weakness, paralysis, and ultimately succumbed to death. However, the underlying mechanism of CV-A6-induced nervous system injury remains elusive. Numerous reports have highlighted the pivotal role of miRNAs in various viral infections.
Methods: Separately established infection and control groups of mice were used to create miRNA profiles of the brain tissues before and after CV-A6 transfection, followed by experimental verification, prediction, and analysis of the results.
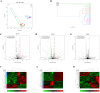
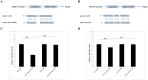
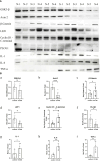
Results: At 2 days post-infection (dpi), 4 dpi, and 2dpi vs 4dpi, we identified 175, 198 and 78 significantly differentially expressed miRNAs respectively using qRT-PCR for validation purposes. Subsequently, we predicted target genes of these differentially expressed miRNAs and determined their potential targets through GO (Gene Ontology) enrichment analysis and KEGG (Kyoto Encyclopedia of Genes and Genomes) enrichment analysis. Finally, we verified the miRNA-mRNA pairing via double luciferase experiments while confirming functional enrichment of target genes through Western Blotting analyses.
Discussion: The results from this study suggest that transcriptional regulation, neuronal necrosis, pro-inflammatory cytokine release, and antiviral immunity are all implicated in the pathogenesis of central nervous system injury in mice infected with CV-A6. Brain injury resulting from CV-A6 infection may involve multiple pathways, including glial cell activation, neuronal necrosis, synaptic destruction, degenerative diseases of the nervous system. It can even encompass destruction of the blood-brain barrier, leading to central nervous system injury. The dysregulated miRNAs and signaling pathways discovered in this study provide valuable insights for further investigations into the pathogenesis of CV-A6.
Keywords: Coxsackievirus A6 (CV-A6); brain; central nervous system; hand foot and mouth disease (HFMD); miRNA.
Copyright © 2024 Sun, Hao, Wu, Qian, Shen and Yu.
Conflict of interest statement
Authors YS, JW, SQ, and SS were employed by the company Wuhan Institute of Biological Products Co. Ltd. The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

